Conserved roles for cytoskeletal components in determining laterality
Version 1 Released on 07 May 2016 under Creative Commons Attribution 4.0 International LicenseAuthors' affiliations
- Department of Biology, School of Arts and Sciences - Tufts University
- Center for Regenerative and Developmental Biology, School of Arts and Sciences - Tufts University
- Department of Biology, Morrissey College of Arts and Sciences - Boston College *. Unregistered author (unverified)
Keywords
Abstract
Consistently-biased left-right (LR) patterning is required for the proper placement of organs including the heart and viscera. The LR axis is especially fascinating as an example of multi-scale pattern formation, since here chiral events at the subcellular level are integrated and amplified into asymmetric transcriptional cascades and ultimately into the anatomical patterning of the entire body. In contrast to the other two body axes, there is considerable controversy about the earliest mechanisms of embryonic laterality. Many molecular components of asymmetry have not been widely tested among phyla with diverse bodyplans, and it is unknown whether parallel (redundant) pathways may exist that could reverse abnormal asymmetry states at specific checkpoints in development. To address conservation of the early steps of LR patterning, we used the Xenopus laevis (frog) embryo to functionally test a number of protein targets known to direct asymmetry in plants, fruit fly, and rodent. Using the same reagents that randomize asymmetry in Arabidopsis, Drosophila, and mouse embryos, we show that manipulation of the microtubule and actin cytoskeleton immediately post-fertilization, but not later, results in laterality defects in Xenopus embryos. Moreover, we observed organ-specific randomization effects and a striking dissociation of organ situs from effects on the expression of left side control genes, which parallel data from Drosophila and mouse. Remarkably, some early manipulations that disrupt laterality of transcriptional asymmetry determinants can be subsequently “rescued” by the embryo, resulting in normal organ situs. These data reveal the existence of novel corrective mechanisms, demonstrate that asymmetric expression of Nodal is not a definitive marker of laterality, and suggest the existence of amplification pathways that connect early cytoskeletal processes to control of organ situs bypassing Nodal. Counter to alternative models of symmetry breaking during neurulation (via ciliary structures absent in many phyla), our data suggest a widely-conserved role for the cytoskeleton in regulating left-right axis formation immediately after fertilization of the egg. The novel mechanisms that rescue organ situs, even after incorrect expression of genes previously considered to be left-side master regulators, suggest LR patterning as a new context in which to explore multi-scale redundancy and integration of patterning from the subcellular structure to the entire bodyplan.
Introduction
Internal organs such as the heart, viscera, and brain display asymmetries in structure and positioning. Loss of correct laterality can result in pathologies contributing to congenital heart disease, a common birth defect [1,2]. Thus, the embryonic origin of consistent laterality is important not only for fundamental questions of evolutionary developmental biology, but also has implications for a broad class of congenital malformations [3–5].
The asymmetrical positioning and orientation of organs is highly conserved [6–9], but despite much recent investigation into the molecular pathways regulating left-right axis formation, there are still many unanswered questions about conservation of early mechanisms [10–12]. One model relies on cilia-driven chiral extracellular fluid flow during neurulation [13]. However, one of the many difficulties with that model as a general answer to the origin of left-right patterning is that numerous phyla establish asymmetry despite lack of cilia, or do so long before cilia are formed [14–17]. This includes single cells [18–21], plants [22,23] , snails [24], nematodes [26–28], fruit flies [29–33], and even amniotes such as chicken [34–36] and pig [37]. A divergent origin of such a fundamental property in even closely related species seems unlikely.
One of the barriers to resolving this problem is that the LR patterning roles of many specific gene products are often investigated in only one model system [38]. Nevertheless, the cytoskeleton is emerging as a component that appears to be relevant to laterality in numerous phyla [23,25,27,39–43]. We previously proposed the model that the intracellular cytoskeleton is an ancient, well-conserved mechanism by which embryos can, at the earliest stages of development, convert chiral molecular structures into true asymmetry of the entire bodyplan [44,6,45,16]. To help unify the data from an evolutionary perspective, and test the opposing predictions of the ciliary vs. intracellular models, we asked: would the same mutations that randomize asymmetry in widely-disparate taxa, including those which do not use cilia, likewise impact asymmetry in the vertebrate Xenopus laevis?
By microinjection of mRNA overexpressing wild type or dominant-negative mutants of cytoskeletal and cytoskeleton-regulating proteins known to regulate asymmetry in very different types of bodyplans, we demonstrate that multiple components of the cytoskeleton are implicated in establishing frog laterality. Here we targeted proteins associated with microtubules and the actomyosin network, including structural proteins ($\alpha$-tubulin), motor proteins (myosins), and moderators of post-translational modification of cytoskeletal components (Mgrn1, Lis1, ect2) with roles in asymmetry identified in Arabidopsis, Drosophila, and mouse. We titered all treatments to produce overall healthy embryos with normal dorsoanterior development and organ morphogenesis, and assayed the expression of laterality markers such as Nodal (Xnr1), Pitx2 and Lefty, and the situs of the heart, gut, and gall bladder. Consistent with a very broad conservation, the same targets were implicated in generating a consistent laterality across the tree of life.
Notably, effects on organ positioning of the gut observed in Drosophila with Myo31DF overexpression [46] are replicated in Xenopus with the frog homologue Myo1D; and the dissociation of left-right organ positioning from Nodal expression observed in mouse mahogunin (Mgrn1) mutants [47] is replicated in Xenopus. We show that some of these steps occur very soon after fertilization and not at later stages (when cilia could be functioning).
This approach also allowed us to address curious discrepancies between molecular marker readouts (of left-side “master regulator” genes) and actual organ situs, which have previously been noted [48]. Indeed it had been proposed that a given species might have multiple overlapping/redundant mechanisms for establishing asymmetry [17], which could have practical consequences for approaches to biomedical disorders of laterality as well as implications for genes like Nodal as left-side “master determinants”. Here, we show that some defects in asymmetric gene expression can be corrected at later points in development to subsequently give correct organ situs. This points to a robust mechanism for symmetry determination that occurs throughout embryogenesis and is not simply determined at a single point.
Together, these data support a unified view of laterality among phyla, identify novel control points for LR patterning during embryogenesis, and suggest LR patterning as a novel paradigm for investigating pattern-correction mechanisms and the interplay of parallel pathways that integrate morphogenesis across levels of organization including the subcellular cytoskeleton, multi-cellular transcriptional domains, and the anatomy of the entire bodyplan.
Experimental
Cloning
Subcloning was carried out into pCS2+ using standard methods. For Mgrn1 clones, EST Biosystems IMAGE:8847517 Xenopus tropicalis clone was sequenced and matched the reference sequence (NM_001016911) exactly. X. laevis cDNA for ect2 (IMAGE:5083828, Thermo Scientific) and X. tropicalis lis1 (IMAGE:5385003), X. laevis myo1d IMAGE:5440331, X. laevis myo1c-b IMAGE:4964888 and X. laevis myo1e.2 IMAGE:4888857 were purchased (Dharmacon). Point mutations were generated using the Agilent QuikChange kit. Ect2-trunc was designed to mimic the Drosophila Pebble splice mutant [49]. Xenopus Flailer [50] was assembled from X. tropicalis gnb5 (IMAGE: 7657559) and X. tropicalis MyoV, (IMAGE:7644816) using the In-Fusion Kit (Clontech). Drosophila myosins were subcloned from constructs received from Kenji Matsuno. All primers used are listed in Supplementary table 6.
Animal Husbandry
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and Tufts IACUC protocol #M2014-79. Xenopus embryos were collected and maintained according to standard protocols [51] in 0.1 x Modified Marc’s Ringers (MMR), pH 7.8, and staged according to [52].
Microinjection
Capped, synthetic mRNAs were dissolved in water and injected into embryos in 3% Ficoll using standard methods [51]. mRNA injections were made into the animal pole of eggs within 30mpf at 14 \textdegree C (unless otherwise stated) using borosilicate glass needles calibrated to deliver a 10 nl injection volume. Embryos injected into targeted blastomeres at the 4-cell stage were selected based on clear differences in pigmentation.
Laterality Assays
Xenopus embryos were analyzed for position (situs) of three organs; the heart, stomach and gallbladder [53] at Stage 45 [52]. Heterotaxic embryos were defined as having a reversal in one or more organs. Only embryos with normal dorsoanterior development and clear left- or right-sided organs were scored. A $\chi^2$ test was used to compare absolute counts of heterotaxic embryos.
In situ hybridization
Whole mount in situ hybridization was optimized using standard protocols [54,55] with probes against Xnr1 (the Xenopus Nodal) [56], Lefty [57] and Pitx2 [58] generated in vitro from linearized template using DIG labeling mix (Roche). A $\chi^2$ test was used to compare absolute counts of embryos with correct (expression on the left lateral plate mesoderm) versus incorrect (absent, bilateral or right-sided) marker expression.
Quantification of plus-end microtubule dynamics
Neural tube explants were dissected from embryos cultured in 0.1 x MMR at 22 \textdegree C to NF Stage (22-24, [52]) and plated onto poly-lysine (100$\mu$g/ml) and laminin-coated (20$\mu$g/ml) coverslips as described previously [59]. Neuronal growth cones were imaged at room temperature 12-18 hours after plating. Live images were collected with a Yokogawa CSU5X1M 5000 spinning disk confocal on a Zeiss Axio Observer inverted motorized microscope with a Zeiss 63X Plan Apo 1.4 NA lens. Images were acquired with a Hamamatsu ORCA R2 CCD camera controlled with Zen software (Zeiss, Thornwood, MY). For time-lapse, images were collected every two seconds for one minute. Laser power for 561nm was 5-15%, with exposure time 600-1000ms. Microtubule dynamics were then quantified using plusTipTracker software [60–62] with MATLAB version 2013a. The same parameters were used for all movies: maximum gap length: 8 frames; minimum track length: 3 frames; search radius range: 5 to 12 pixels; maximum forward angle: 50 \textdegree; maximum backward angle: 10 \textdegree; maximum shrinkage factor: 0.8; fluctuation radius: 2.5 pixels. Only cells with a minimum number of 10 track events in a movie were included for analysis.
Results
Microinjection alone of Xenopus embryos immediately post-fertilization does not disrupt laterality.
To determine if the act of microinjection itself, or of a large amount of any mRNA, into the animal pole immediately post-fertilization is sufficient to disrupt laterality, Xenopus laevis embryos were microinjected with either water or $\beta$-galactosidase mRNA within 30 minutes of fertilization (mpf). Embryos at stage 45 were scored for the positioning of the heart, gall bladder and stomach (Fig. 1A). No defects in positioning of the visceral organs were observed (Fig. 1B); $\beta$-galactosidase signal was observed by the initiation of the first cell division (data not shown), as previously demonstrated for cofilin-Tomato [45]. Likewise, embryos injected with water were scored for the laterality of Nodal (Xnr1) at neurula stages (Fig. 1C). No alterations to the laterality of Xnr1 laterality were observed (Fig. 1D). Therefore, neither microinjection per se, nor the injection of large amounts of mRNA soon after fertilization, represents an insult sufficient to disrupt laterality.
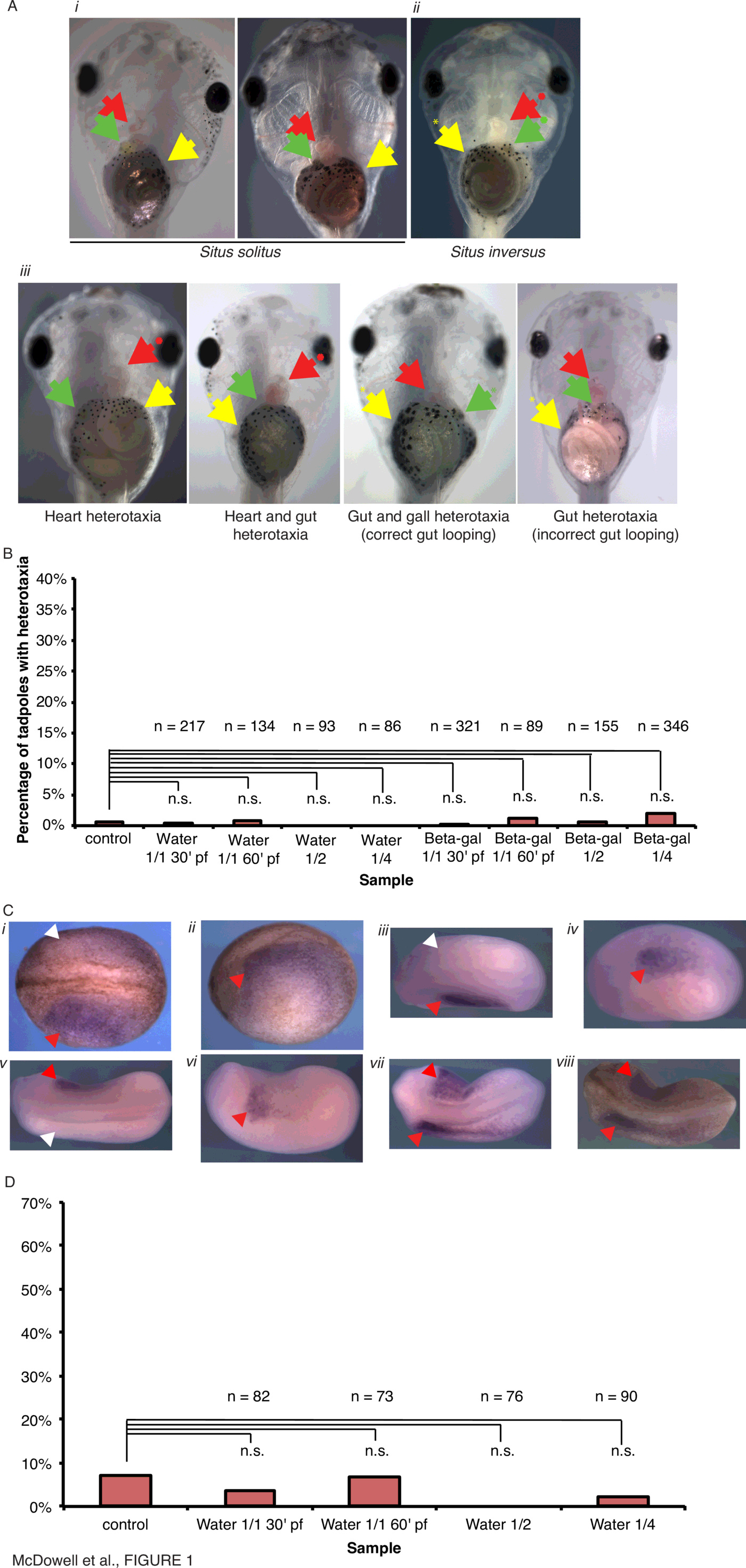 | Figure 1 Figure 1. Injection of water or high amounts of $\beta$-galactosidase mRNA does not result in laterality defects in Xenopus laevis. (A) Positioning of visceral organs at Stage 45 in Xenopus laevis. (i) Wild-type (situs solitus) embryos, ventral view, showing the normal arrangement of the stomach (yellow arrowhead), heart apex (red arrowhead), and gall bladder (green arrowhead). (ii) An heterotaxic embryo (ventral view) showing reversal of all three organs, i.e., situs inversus. (iii) Heterotaxic embryos (ventral view) showing various incorrect organ situs. (B) Embryos were injected into the animal pole with 10 nl water or mRNA encoding $\beta$-galactosidase at 1.5ng nl$^{-1}$ at various timepoints and scored for visceral organ situ. (C) Examples of Xnr1 expression assayed by in situ hybridization with absence of Xnr1 expression indicated with white arrowhead, and presence of Xnr1 indicated by red arrowheads: (i) correct left-sided Xnr1 expression, dorsal view, and (ii) left lateral view, unbleached embryo; (iii) correct left-sided Xnr1 expression, dorsal view, and (iv) left lateral view, bleached embryo; (v) incorrect right-sided Xnr1 expression, dorsal view, and (vi) right lateral view, bleached embryo; (vii) incorrect bilateral Xnr1 expression, dorsal view, bleached and (viii) unbleached embryos. (D) Embryos were injected into the animal pole with 10 nl water at various timepoints and scored for laterality of Xnr1 expression. |
Disrupting microtubule architecture affects laterality.
$\alpha$-tubulin and Tubgcp2, a protein of the $\gamma$-tubulin complex, are components of the microtubule cytoskeleton that affect symmetry of axial organs in Arabidopsis [39,63,64,41,23] and embryonic laterality in Xenopus (45). To determine if a mutant form of $\alpha$-tubulin corresponding to a right-helical mutant which skews the direction of root growth [64] has a similar effect on vertebrate laterality as other $\alpha$-tubulin mutants identified in Arabidopsis [45], Xenopus laevis embryos were microinjected with mRNA encoding a mutant form of Xenopus $\alpha$-tubulin, the T56I mutant (Fig. 2, Supplementary Tables 1, 2, Supplementary Fig. 1).
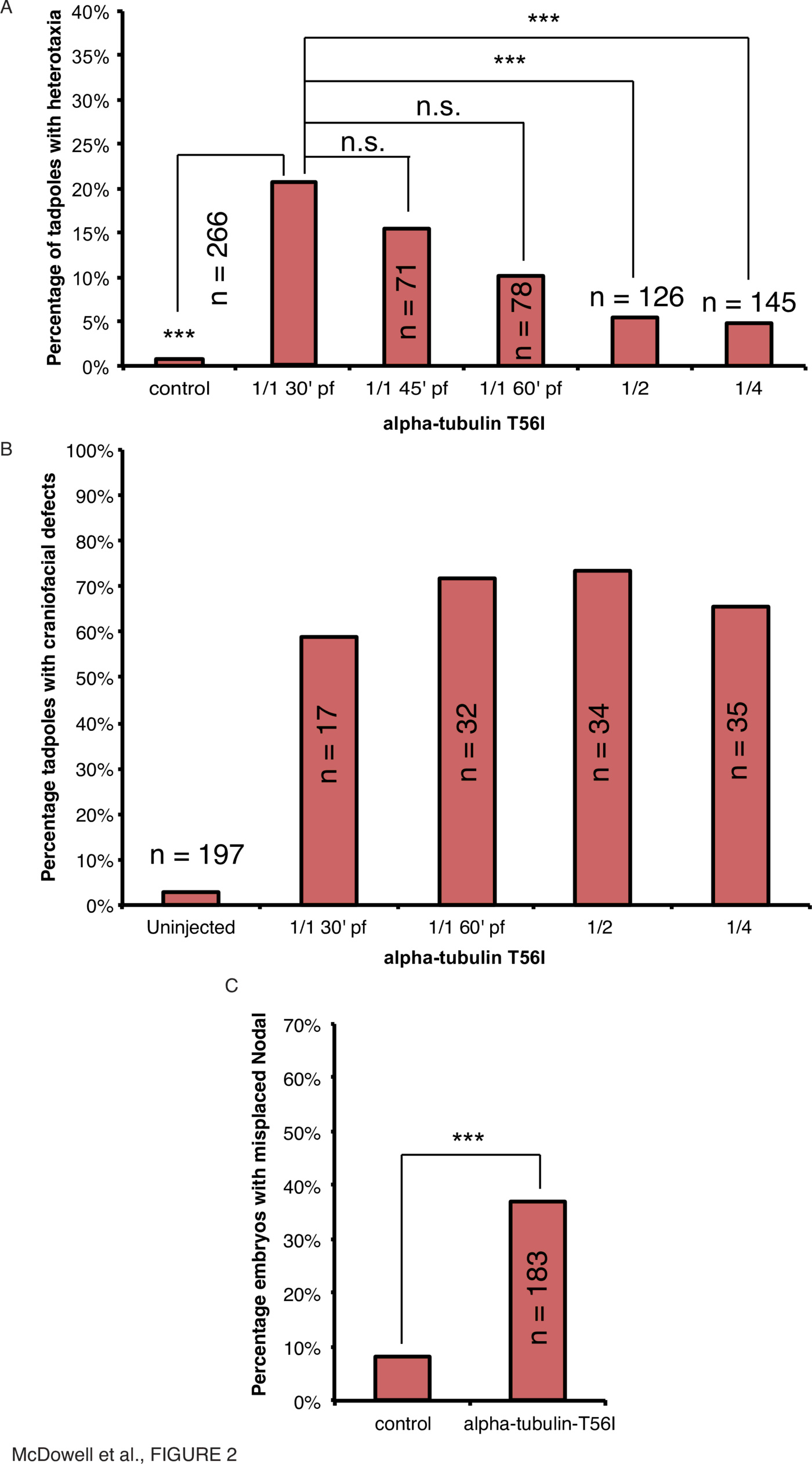
Craniofacial abnormalities were also present in $\alpha$-tubulin-T56I-injected tadpoles (Fig. 2B). In contrast to the decrease in heterotaxia seen with later injection, craniofacial defects occurred consistently in 1-cell, 2-cell and 4-cell injections in more than 50% of tadpoles (Fig. 2B). This serves as a convenient internal positive control for the activity of the mutant tubulin protein in development. Overall, these data illustrate that the effect of $\alpha$-tubulin-T56I on heterotaxia is required in the earliest timeframe of development for left-right patterning, and not at a later symmetry-breaking event, as later processes involved in craniofacial development are still affected equally regardless of the injection timepoint.
To place this mechanism within the known major steps of left-right patterning, we asked whether introduction of the tubulin mutant would affect the sidedness of the canonical left-side marker Xnr1 [65,66]. Injection of $\alpha$-tubulin-T56I mRNA within 30mpf resulted in incorrect (right-sided, bilateral or absent) expression of Xnr1 in 37% of neurula-stage embryos compared to 7% in uninjected controls (Fig. 2C, $\chi^2<0.001, n=183$). Therefore, transcriptional control points upstream of organ orientation are also affected by $\alpha$-tubulin-T56I. Taken together, our data show that a tubulin mutation that regulates chirality in plants likewise randomizes organ laterality in Xenopus embryos. Moreover, the introduction of these dominant-negative mutations are only effective when performed immediately after fertilization, revealing that any tubulin-derived structures must be acting in LR patterning at very early stages of development.
Disrupting microtubule regulation by post-translational modification affects laterality.
Tubulins are modified by a variety of post-translational modifications, such as acetylation, phosphorylation and ubiquitylation [67]. The process of ubiquitylation, whereby a ubiquitin moiety is covalently fused to a protein through an electron-rich group (such as internal lysine, cysteine, serine or threonine residues, or the N-terminal amino group [68]), can target the protein for localization to a different part of the cell or for degradation using various combinations of mono- or polyubiquitylation [69]. The E3 ligase, Mahogunin ring-finger 1 (Mgrn1), polyubiquitylates $\alpha$-tubulin to target it for degradation [70,71]. Mgrn1-null mouse mutants exhibit phenotypes such as late-onset spongiform neurodegeneration and laterality defects such as congenital heart defects and situs inversus [47,72]. In particular, the effect of Mgrn1 on left-right patterning was found to be uncoupled from the expression of Nodal and yet it affected laterality of downstream Nodal targets, leading the authors to conclude that Mgrn1 and tubulin ubiquitylation had a novel role in early LR patterning [47].
To investigate whether the effects of Mgrn1 on laterality observed in mouse were conserved in frog, Xenopus laevis embryos were microinjected with mRNA encoding Xenopus laevis Mgrn1, or the mutant forms Mgrn1-G2A (a non-myristoylatable mutant which alters the strength of its interaction with membranes [73]) or Mgrn1-C314D (the homologue of the C316D catalytically inactive mutant [70]), at various timepoints and scored for organ situs and Xnr1 laterality (Fig. 3, Supplementary Tables 1, 2, Supplementary Fig. 1).
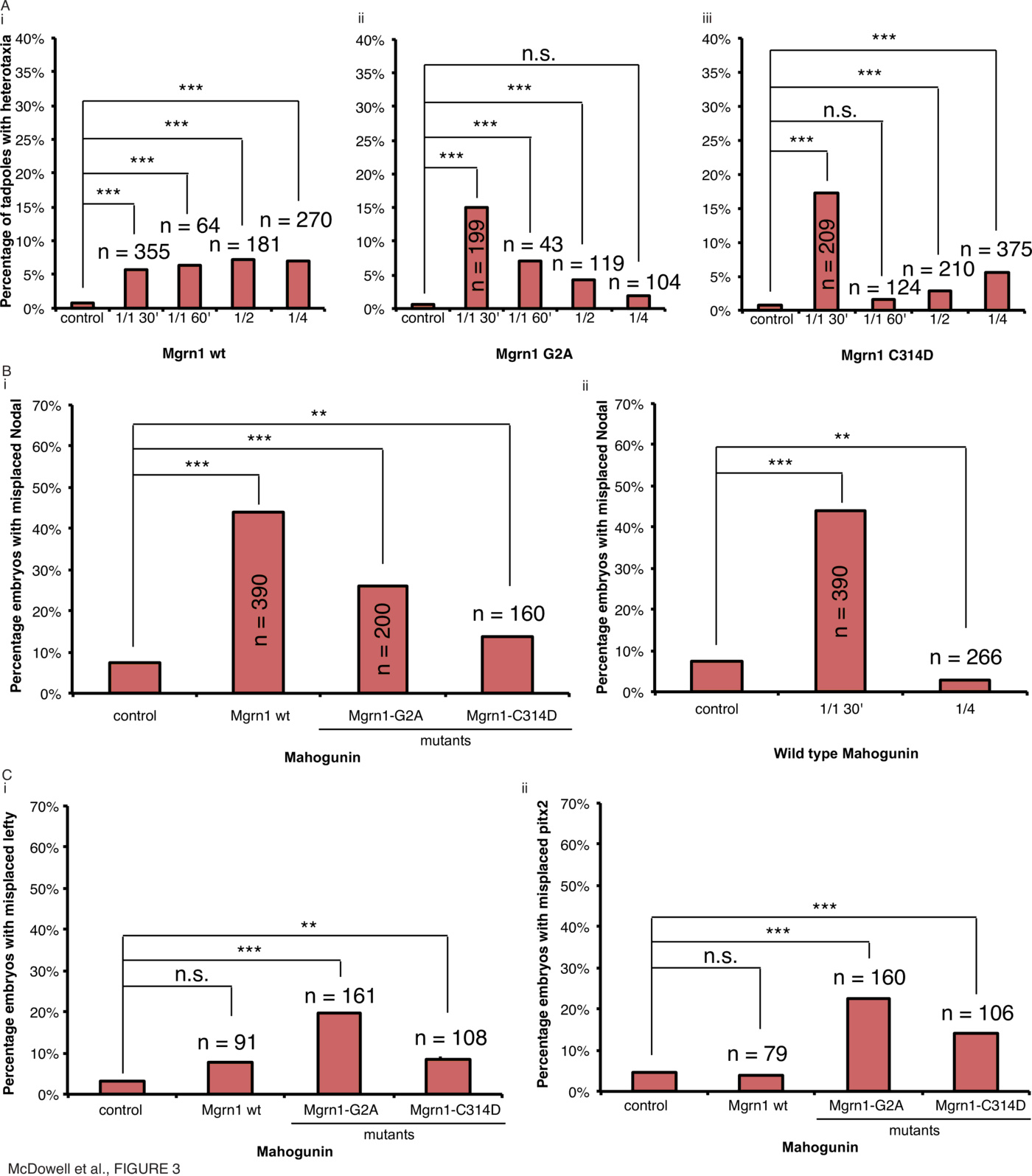
Injection of wild type Mgrn1 mRNA at various timepoints induced heterotaxia in only 6% of tadpoles at 30 minutes (Fig. 3A(i), $\chi^2<0.001, n=355$), 6% of tadpoles at 60 minutes (Fig. 3B(i), $\chi^2<0.001, n=64$), 7% of tadpoles in 1 of 2 cells (Fig. 3A(i), $\chi^2<0.001, n=181$), and 7% of tadpoles in 1 of 4 cells (Fig. 3A(i), $\chi^2<0.001, n=270$), showing no effect at any stage of development due to overexpression of the wild type mahogunin protein as the rate of heterotaxia is below our biologically-relevant threshold of 10%. In contrast, injection of Mgrn1G2A mRNA within 30mpf resulted in 15% organ heterotaxia (Fig. 3A(ii), $\chi^2<0.001, n=199$). Notably, the efficacy dropped with increasing developmental timepoint of injection, with only 7% of tadpoles displaying heterotaxia when injected at 60mpf (Fig. 3A(ii), $\chi^2<0.001, n=43$), 4% of tadpoles displaying heterotaxia at 1 of 2 cell (Fig. 3A(ii), $\chi^2<0.001, n=119$) and 2% of tadpoles displaying heterotaxia at 1 of 4 cell (Fig. 3A(ii), $\chi^2=0.143, n=104$). Likewise, injection of Mgrn1-C314D mRNA within 30mpf resulted in organ heterotaxia in 17% of tadpoles (Fig. 3A(iii), $\chi^2<0.001, n=209$); this effect also dropped to 2% at 60 minutes (Fig. 3A(iii), $\chi^2=0.236, n=124$), 3% at 1 of 2 cell (Fig. 3A(iii), $\chi^2<0.001, n=210$) and 6% at 1 of 4 cell (Fig. 3A(iii), $\chi^2<0.001, n=375$). Therefore, whilst overexpression of the wild type protein has very little effect on organ positioning, overexpression of both the G2A and C314D mutants result in randomization of organ situs, but only when the mRNA is injected within the first 30mpf.
Mgrn1 injections dissociate organ situs from sidedness of asymmetric gene expression
To assess whether the effects on organ situs were preceded by alterations in expression of known markers of embryonic laterality, the sidedness of Xnr1 expression in embryos with modified Mgrn1 function was assessed by in situ hybridization (Fig. 3B). Injection of wild type Mgrn1 mRNA within 30mpf resulted in incorrect expression of Xnr1 in 44% of neurula-stage embryos compared to 8% in uninjected controls (Fig. 3B(i), $\chi^2<0.001, n=390$). Injection of Mgrn1-G2A mRNA resulted in incorrect expression in 26% of embryos (Fig. 3B(i), $\chi^2<0.001, n=200$) and injection of Mgrn1-C314D mRNA resulted in incorrect expression in 14% of embryos (Fig. 3B(i), $\chi^2=0.009, n=160$). Therefore all Mgrn1 proteins have an effect on the positioning of Xnr1 expression but surprisingly the effect of wild type Mgrn1 on Xnr1 does not translate into an effect on organ situs: far more embryos had abnormal Xnr1 expression than had reversals of organ situs. We interpret these results to indicate the existence of a mechanism that can correct abnormal laterality subsequent to Xnr1 expression.
To determine if this effect on the laterality of Xnr1 expression has a temporal dependence, embryos were injected within 30 minutes, or into 1 of 4 cells, with wild type Mgrn1 mRNA and the laterality of Xnr1 expression was assessed by in situ hybridization (Fig. 3B(ii)). Injection of wild type Mgrn1 mRNA within 30mpf resulted in incorrect expression of Xnr1 in 44% of neurula-stage embryos compared to 7% in uninjected controls (Fig. 3B(i), $\chi^2<0.001, n=390$) whereas injection into 1 of 4 cells resulted in incorrect expression in only 3% of embryos (Fig. 3B(ii), $\chi^2=0.007, n=266$). Therefore the sidedness of Xnr1 expression also demonstrates a temporal dependence in a similar manner to organ heterotaxia: only extremely early manipulations of this pathway are sufficient to randomize asymmetric gene expression – waiting even an hour later after fertilization (still long before cilia appear) is not effective in perturbing left-right patterning.
Expression of Xnr1 is followed at later stages by the expression of Lefty [74] and Pitx2 [75], and we next asked where in this pathway the cytoskeletal protein (and the corrective mechanisms we uncovered) might act. To assess whether the misexpression of Xnr1 caused by Mgrn1 overexpression also resulted in misexpression of Lefty and Pitx2, embryos were injected and the laterality of Lefty and Pitx2 expression was assessed by in situ hybridization (Fig. 3C, Supplementary Tables 3, 4). Injection of wild type Mgrn1 mRNA within 30mpf resulted in incorrect expression of Lefty in 8% of neurula-stage embryos compared to 3% in uninjected controls (Fig. 3C(i), $\chi^2=0.014, n=91$) and incorrect expression of Pitx2 in 4% of neurula-stage embryos compared to 4% in uninjected controls (Fig. 3C(ii), $\chi^2=0.782, n=79$). Injection of Mgrn1-G2A mRNA resulted in incorrect expression of Lefty in 20% of neurula-stage embryos compared to 3% in uninjected controls (Fig. 3C(i), $\chi^2<0.001, n=161$) and incorrect expression of Pitx2 in 23% of neurula-stage embryos compared to 4% in uninjected controls (Fig. 3C(ii), $\chi^2<0.001, n=160$). Injection of Mgrn1-C314D mRNA resulted in incorrect expression of Lefty in 8% of neurula-stage embryos compared to 3% in uninjected controls (Fig. 3C(i), $\chi^2<0.001, n=108$) and incorrect expression of Pitx2 in 14% of neurula-stage embryos compared to 4% in uninjected controls (Fig. 3C(ii), $\chi^2<0.001, n=106$). Therefore, the randomization of Xnr1 expression induced by wild type Mgrn1 overexpression is corrected by the time of Lefty and Pitx2 expression. Interestingly, Mgrn1-C314D does not have a large effect on misexpression of early laterality markers but does induce a change in organ situs, suggesting the existence of a pathway leading from early cytoskeletal events to organ positioning that is parallel to the canonical Nodal-Lefty-Pitx2 cascade. Only Mgrn1-G2A has a consistent effect on laterality throughout development looking at all markers of laterality evaluated here. This uncoupling of the expression of Xnr1 and its effect on downstream targets is consistent with findings on Nodal in Mgrn1 regulation in mice [47], and is the first example of mechanistic conservation of a cytoskeletal asymmetry component between Xenopus and mammalian embryos.
Laterality defects caused by disruption of transport along microtubules are not specific to early cytoskeletal rearrangements.
Movement of cargos along microtubules is carried out by motor proteins such as dyneins, which are minus-end-directed motor proteins and have been shown to be important for laterality, in particular left-right dynein, which has ciliary [76,77] and non-ciliary roles [78,79,6,80]. The dynein accessory factor Lis1 is required for dynein-dependent processes such as positioning of the nucleus during neuronal migration in humans [81] and spermatogenesis in Drosophila [82], and in particular has a role in asymmetric cell division demonstrated in mouse [83].
Lis1 is a component of the dynein/dynactin motor complex, regulating trafficking and being responsible for the smooth-brain phenotype of lissencephaly. It is also required for the organization of non-centrosomal cortical microtubules [84] and has been implicated in planar cell polarity [85] and asymmetric cell division [83]. In the regulation of dyneins and the dynamics of microtubules, Lis1 complexes with 14-3-3 [86], which has also demonstrated a role in regulating left-right asymmetry [87]. Lis1-N99 acts by forming non-functional heterodimers with Lis1, whereas Lis1-C137 competes with Lis1 for cofactor binding, and both mutants are associated with mitotic catastrophe and cell death [88].
To determine if forms of Lis1 with dominant-negative effects in human cell lines [88] function in vertebrate LR asymmetry, embryos were microinjected with mRNA encoding mutant forms of Xenopus Lis1, the N99 and C137 mutants [88], within 30mpf and scored for organ situs and Xnr1 laterality (Fig. 4, Supplementary Tables 1, 2, Supplementary Fig. 1).
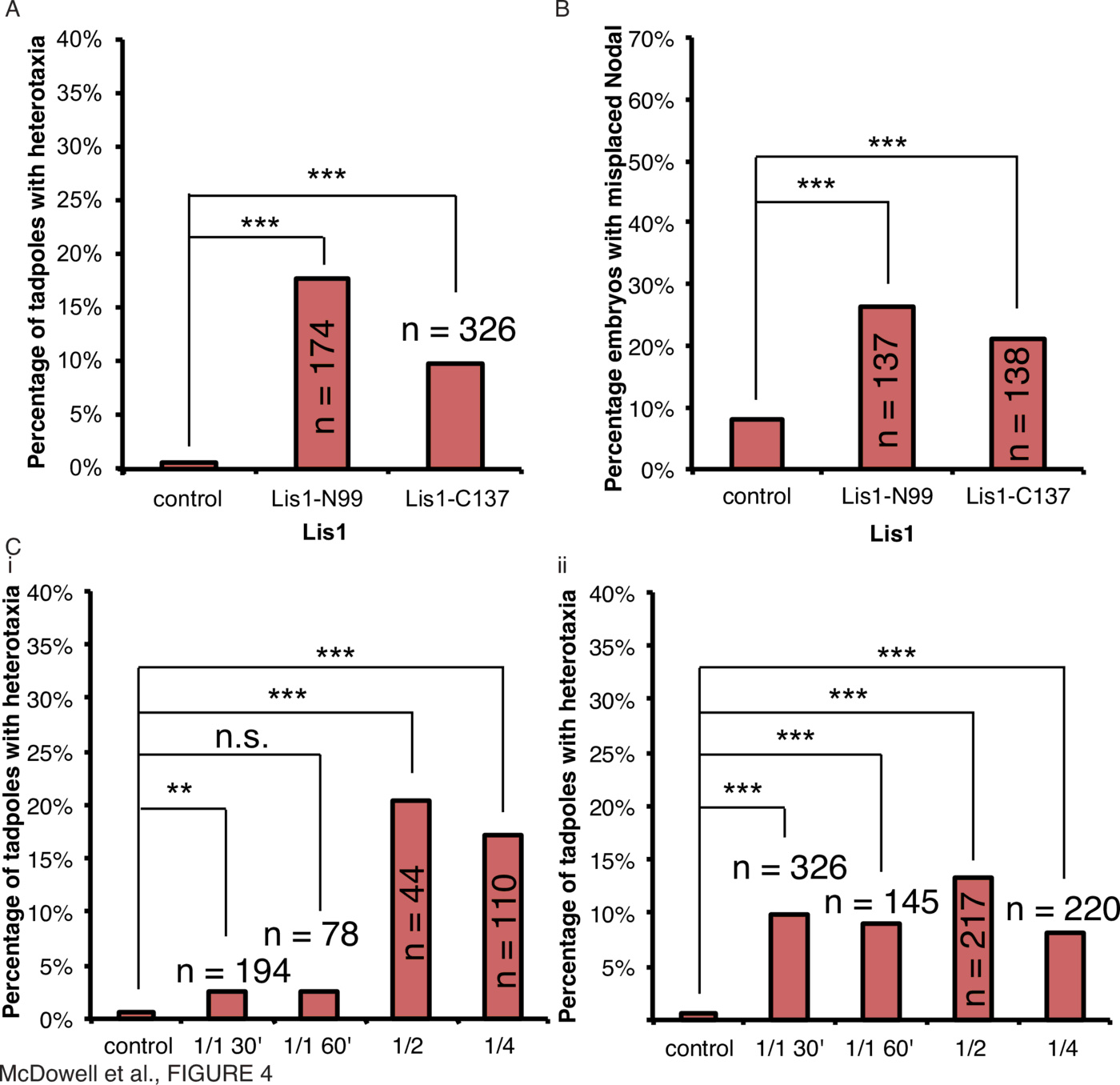
To assess whether Lis1 mutants act as regulators of early cytoskeletal mechanisms affecting laterality in a manner similar to $\alpha$-tubulin and mahogunin above, injections were carried out at various timepoints for both Lis1-N99 (Fig. 4C(i)) and Lis1-C137 (Fig. 4C(ii)). Strikingly, injections of 12ng Lis1-N99 were toxic when injected after 30mpf. Injection of $\ge$4ng Lis1-N99 mRNA within 30mpf induced heterotaxia in 3% of tadpoles (Fig. 4C(i), $\chi^2=0.002, n=194$), injection 60mpf induced heterotaxia in 3% of tadpoles (Fig. 4C(i), $\chi^2=0.052, n=78$), into 1 of 2 cells induced heterotaxia in 20% of tadpoles (Fig. 4C(i), $\chi^2<0.001, n=44$), and into 1 of 4 cells induced heterotaxia in 17% of tadpoles (Fig. 4C(i), $\chi^2<0.001, n=110$). Unlike the proteins tested above, Lis1 can randomize at both early and later stages. Likewise, injection of $\ge$2ng Lis1-C137 mRNA within 30mpf resulted in organ heterotaxia in 10% of tadpoles (Fig. 4A, C(ii), $\chi^2<0.001, n=326$), injection 60mpf induced heterotaxia in 9% of tadpoles (Fig. 4C(ii), $\chi^2<0.001, n=145$), into 1 of 2 cells induced heterotaxia in 13% of tadpoles (Fig. 4C(ii), $\chi^2<0.001, n=217$), and into 1 of 4 cells induced heterotaxia in 8% of tadpoles (Fig. 4C(ii), $\chi^2<0.001, n=220$).
These data suggest that intracellular microtubule-dependent trafficking and motor protein-dependent cargo movement machinery is important for normal laterality. The degree of randomization is also consistent regardless of the time of embryo injection, providing a contrast to other results.
Laterality defects caused by disruption of regulation of the actomyosin complex by the Rho-GEF ect2 are not specific to early cytoskeletal rearrangements.
Alongside microtubule assemblies, the cytoskeleton consists of actin filaments, which, with the myosin motor proteins, comprise the actomyosin machinery. Pebble is a Rho-GEF which regulates actomyosin complexes (89) and is involved in cytokinesis in Drosophila and in particular regulates the separation of syncytial nuclei in the Drosophila embryo [89]; it was recently shown to be important in Drosophila hindgut rotation through laterality defects arising from mutant forms of the protein [49]. Pebble is homologous to ect2 in vertebrates and we constructed a truncation mutant of the Xenopus form, ect2-trunc, to investigate the effect of this protein on laterality. To determine if the ect2 truncation mutant would have a similar effect on laterality in Xenopus as it does in Drosophila, embryos were microinjected with mRNA encoding a mutant, Xenopus ect2-trunc, and scored for organ situs and the laterality of Xnr1, Lefty and Pitx2 expression (Fig. 5, Supplementary Tables 1, 2, 3, 4, Supplementary Fig. 1).
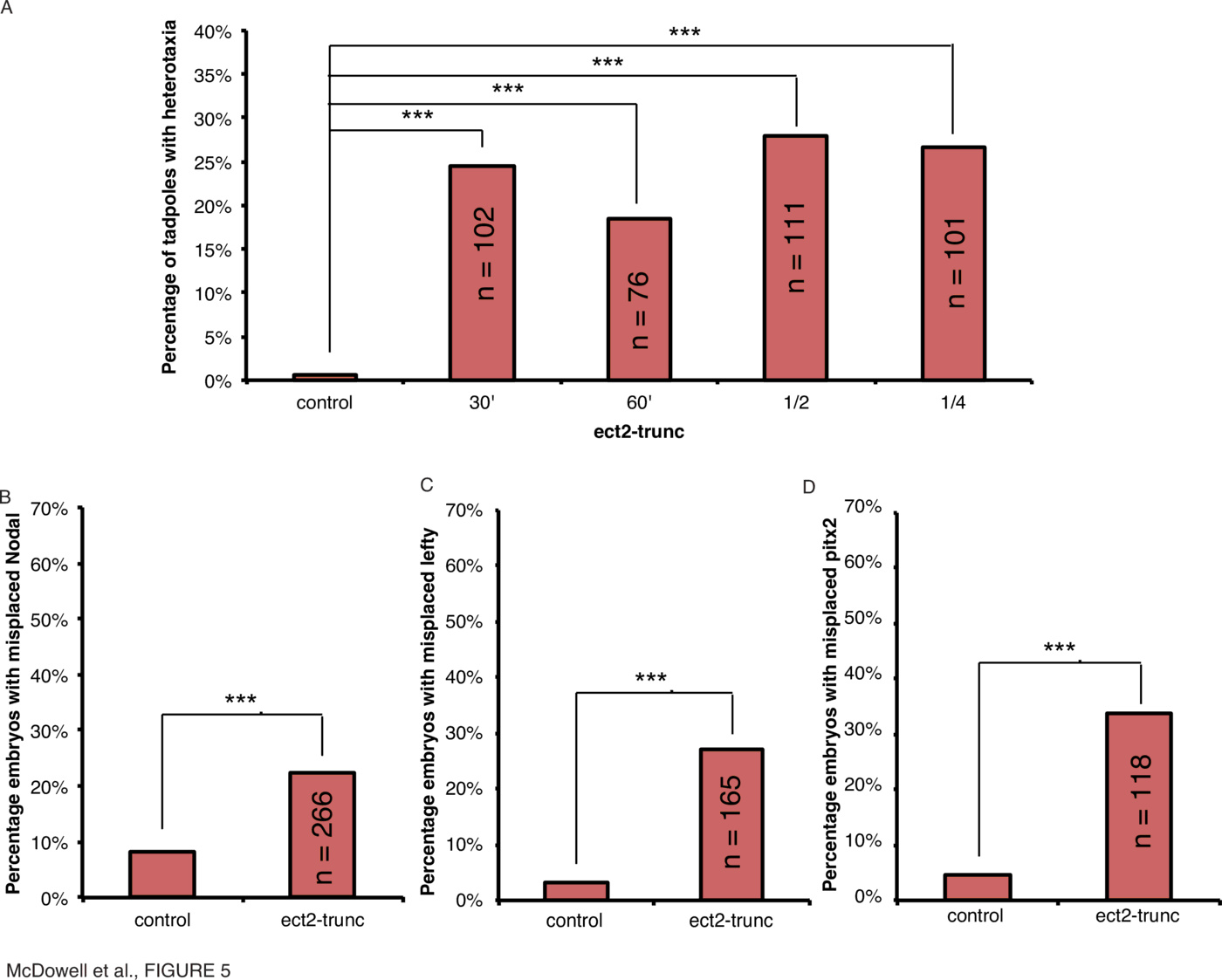
Disruption of the myosins immediately post-fertilization affects laterality.
As the motor protein component of the actomyosin cytoskeleton, myosins are important for transporting cargo throughout the cell. Myosins have also been identified in Drosophila as having roles in asymmetric hindgut rotation [30,43]. Myo31DF is involved in left-right patterning in Drosophila [43,90], and Myo61F overexpression antagonizes Myo31DF's function, leading to inversion of the gut [32,90].
Myosins have also demonstrated a role in transporting ion channels, which play important roles in LR patterning [17]. Ion channel isoforms are transported by myosins such as myosin V [91]. A mouse line bearing a mutant form of myosin Va is named Flailer due its effect on Purkinje cells [50]. The protein is unable to bind to actin filaments but still binds organelles through its globular tail domains, and acts as a dominant-negative by competing with wild-type Myosin VA [50]. To determine if a myosin implicated in ion channel transport was important for regulating early laterality determination, embryos were microinjected with mRNA encoding the Xenopus form of the Flailer mutant at different timepoints of development and scored for organ situs and Xnr1 expression laterality (Fig. 6, Supplementary Tables 1, 2, Supplementary Fig. 1).
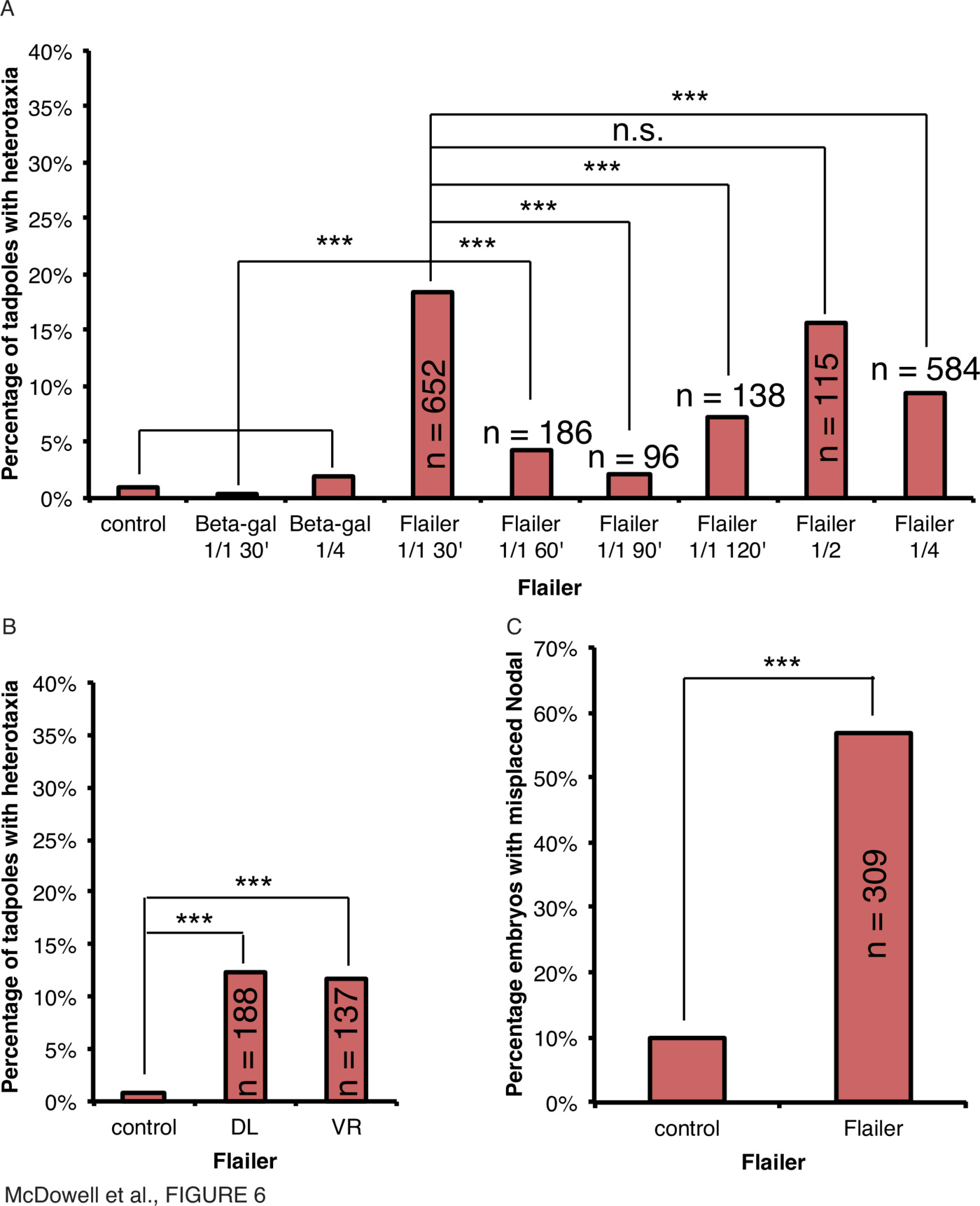
To determine if the results of injections at the 4-cell stage could be explained by disturbances of ciliary flow at the gastrocoel roof plate (GRP), Flailer mRNA was microinjected into the dorsal left (DL) or ventral right (VR) blastomeres (Fig. 6B). Organ heterotaxia was observed at a rate of 12% in both DL- and VR- injected embryos compared to 1% in uninjected embryos (Fig. 6B, $\chi^2<0.001, n=188$ (DL); 137 (VR)), demonstrating that even though heterotaxia was observed, there was no difference between blastomeres that are or are not necessary for asymmetric ciliary flow signaling.
Injection of $\ge$250pg Flailer mRNA within 30mpf resulted in incorrect expression of Xnr1 in 57% of neurula-stage embryos compared to 7% in uninjected controls (Fig. 6C, $\chi^2<0.001, n=309$). Therefore laterality signals upstream of organ orientation are also affected by disruption by the mutant Flailer, identified in mouse, immediately after fertilization.
Next, myosin mutants implicated in laterality defects in Drosophila were microinjected into embryos of Xenopus laevis as mRNA encoding Drosophila forms of Myo31DF or Myo61F, or Xenopus laevis forms, Myosin1d (corresponding to Myo31DF), Myosin1cA or Myosin 1e2 (both corresponding to Myo61F), within 30mpf. Embryos were stored at 14 {\textdegree}C during early cleavage stages and developed at 18 {\textdegree}C to Stage 45 to score the positioning of the visceral organs or to neurula stages for in situ hybridization using a probe against Xnr1 and scoring laterality of Xnr1 expression (Fig. 7, Supplementary Tables 1, 2, Supplementary Fig. 1).
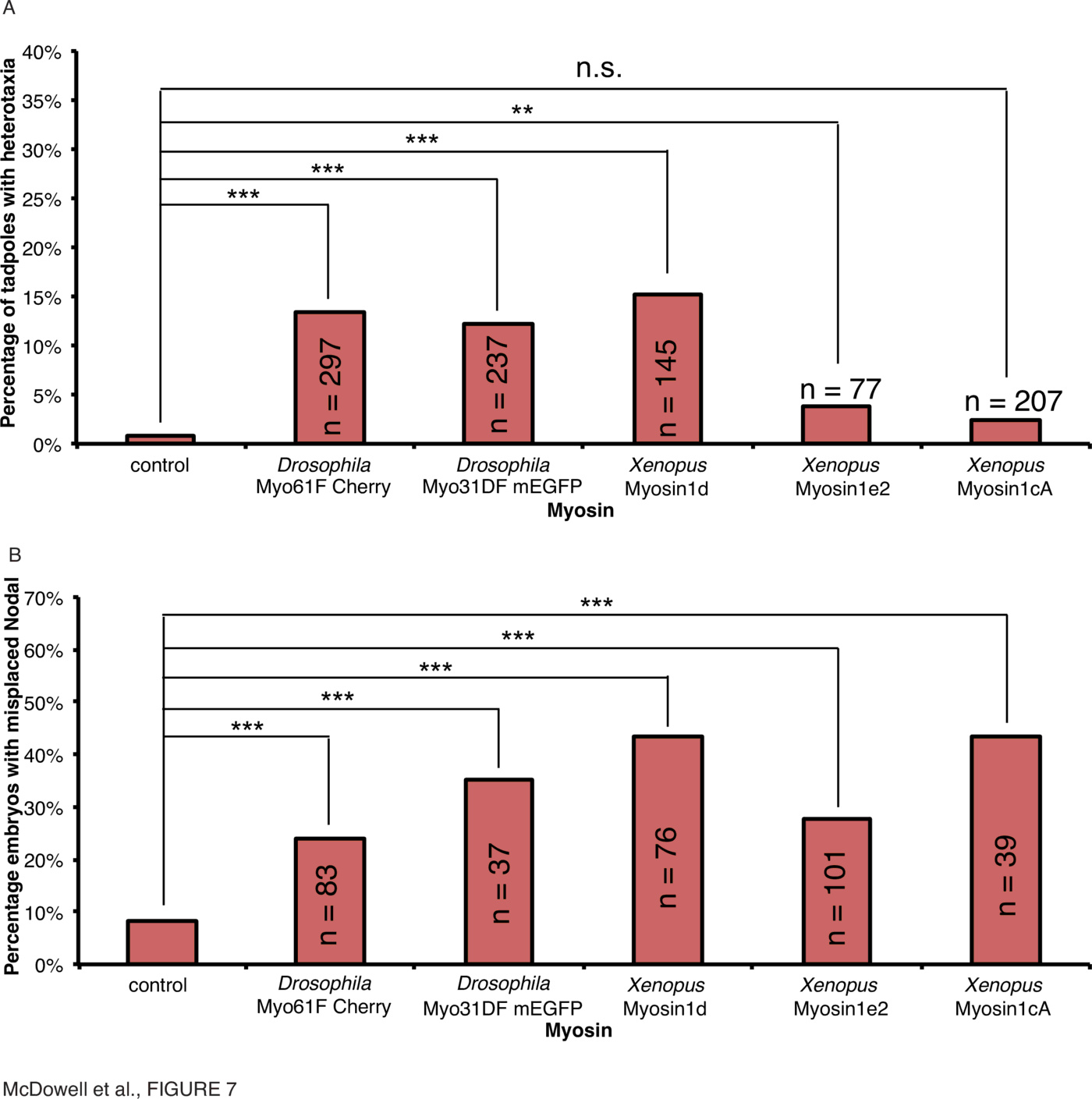
Myo31DF and Myo61F are implicated in gut and genitalia asymmetry in Drosophila [43,90], and Myo61F overexpression antagonizes Myo31DF's function [32,90]. Overexpression of both Drosophila proteins in Xenopus results in the observation of heterotaxia in various organs. Most striking is that only recently were the tissue-specific functions of these proteins identified in Drosophila, and it appears that Myo61F is important in genitalia-turning, whereas Myo31DF is involved in the stomach [46]. Myosin1d, the homologue of Myo31DF, affects stomach laterality in all observed cases of heterotaxia (Supplementary Fig. 1), but the frog homologues of Myo61F apparently have no effect. As genitalia do not turn in Xenopus, this suggests that perhaps homologues of Myo61F do not exert an effect on organ patterning in frog, but do in Drosophila.
Injection of Myo61F ($\ge$1ng), Myo31DF ($\ge$7.5ng), myosin1d ($\ge$5ng), myosin1cA ($\ge$5ng) and myosin1e2 ($\ge$5ng) mRNA within 30mpf resulted in incorrect expression of Xnr1 in 35% (Fig. 7B, $\chi^2<0.001, n=37$), 24% (Fig. 7B, $\chi^2<0.001, n=83$), 43% (Fig. 7B, $\chi^2<0.001, n=76$), 44% (Fig. 7B, $\chi^2<0.001, n=39$) and 28% (Fig. 7B, $\chi^2<0.001, n=101$) of neurula-stage embryos compared to 7% in uninjected controls, respectively. Therefore laterality signals upstream of organ orientation are also affected by overexpression of both the Drosophila myosins and their frog homologues, immediately after fertilization. The discrepancy between the effect of Xenopus myosin1cA and myosin1e2 on organ situs and Xnr1 expression illustrates, as with Mgrn1, an important dissociation of the effect of Nodal expression and the patterning of visceral organs.
Taken together, our data indicate that the myosins are involved in laterality in frog embryos as well as in Drosophila. However, we note that whilst all myosins affect the laterality of Xnr1 expression, the effects on organ situs are diverse, and in particular the comparison with the roles of the homologous proteins in Drosophila shows that the situation in fly is mirrored in frog in the appropriate context of gut turning.
Microtubule dynamics are affected by tubulin mutants effecting chirality in the early embryo.
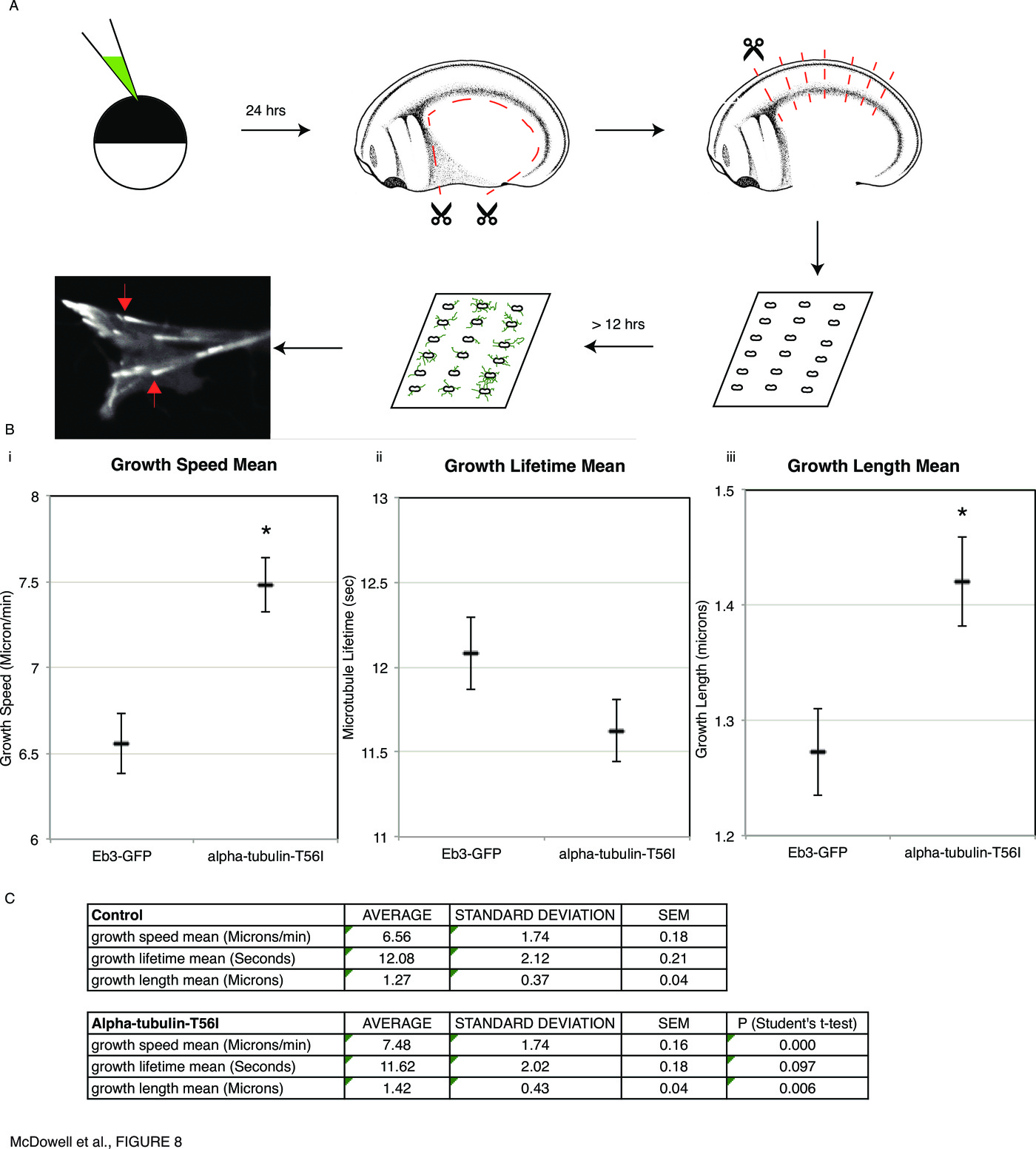
What properties of the cytoskeleton might be altered by the misexpression of mutants known to randomize asymmetry? To investigate the effectors of chirality at the cellular level, we chose to quantify microtubule plus-end dynamics [92] in explanted Xenopus neuronal growth cones [59] by coinjection of $\alpha$-tubulin-T56I and GFP-tagged End-binding protein Eb3, followed by quantitative analysis of microtubule plus-end dynamics using plusTipTracker software ([62], Fig. 8A).
Discussion
We show that an assortment of proteins, implicated in laterality regulation in a wide range of phyla, is capable of specifically randomizing left-right asymmetry in Xenopus embryos manipulated immediately post-fertilization. Neither water nor large amounts of non-specific mRNA affect laterality when injected immediately post-fertilization, demonstrating that microinjection per se at this early stage does not disrupt the cytoskeleton enough to affect asymmetry: the LR axis is not a highly labile system perturbed by nonspecific manipulations. Thus, the early embryo is a robust, specific assay for components required for normal LR patterning.
In subsequent experiments, we scored the positioning of the 3 asymmetric organs and reported as percent heterotaxic the number of animals exhibiting aberrant situs (but normal morphology) of any of the three. We titered all reagents to levels that produced normal dorsoanterior index (DAI) phenotypes and organ morphogenesis, so that LR reversals can be interpreted cleanly without confounding effects of non-specific toxicity (and, the differences in results reported among various treatments and assays (organs vs. gene expression) were not due to differential survival). In interpreting the heterotaxia incidence percentages resulting from any of the reagents, it should be noted that the absolute highest heterotaxia level that can be observed is 87.5%: even if each organ is fully (independently) randomized, in a small percentage (12.5%) of the cases, all three organs will by chance land on their normal sides, appearing as though the animal had wild-type situs. Thus, all percentages of organ heterotaxia are relative to a ceiling that is not 100% but 87.5%.
The Cytoskeleton: a common factor linking LR patterning across widely-disparate phyla
We have identified a conserved role for the cytoskeleton in Xenopus immediately post-fertilization in affecting left-right patterning during embryogenesis. Proteins and their mutations identified across kingdoms of life, from plants such as Arabidopsis, to invertebrates such as Drosophila, and to vertebrate mammals such as mouse, have all been shown to replicate their effects on laterality, and in some cases organ- and Nodal-specific phenotypes, in the frog Xenopus laevis. Not only is the catalogue of proteins tested capable of affecting laterality immediately post-fertilization, but individual proteins demonstrate subtle characteristics that demonstrate, and some cases replicate what is observed in other organisms, their different roles in regulating the cytoskeletal role in left-right patterning. For example, the injection of mutants of Mgrn1 demonstrated their role in affecting organ situs more than overexpression of the wild-type protein. Conversely, overexpression of wild type protein affects laterality of Xnr1 expression more than the mutant forms (Fig. 3). This matches observations of the dissociation of Nodal expression from downstream laterality in mouse [47]. Especially striking is that wild-type Mgrn1 randomizes Xnr1 expression quite effectively but has almost no effect at all on organ situs. These are the first data showing a common cytoskeletal protein involved in Xenopus and mouse. Similarly, the ect2 truncation mutant induced heterotaxies mostly in the gut, replicating observations for the role of the homologue Pebble in Drosophila [49]. Observations of the Class I myosins from Drosophila, Myo31DF and Myo61F, were also not only confirmed in Xenopus, but the use of the Xenopus homologue to Myo31DF, Myo1d, matched exactly the role for Myo31DF in gut turning in Drosophila [46], whereas Myo61F, required in turning of Drosophila genitalia [46], was less specific in the organ situs it affected, and the Xenopus homologues could not affect organ situs but did affect the laterality of Xnr1 expression (Fig. 7). This is, to our knowledge, the first demonstration of a laterality pathway component in common between Drosophila and vertebrates [30]. It is striking that LR phenotypes in Drosophila and mouse are so accurately replicated in the Xenopus model. Future work will continue the effort of testing molecular components in diverse model species, and reveal mechanisms by which the same very early asymmetry-generating machinery is exploited by highly diverse bodyplans.
Asymmetrical positioning of organs is highly conserved [6–8], but a conserved mechanism for laterality is still under much discussion [10–12]. One class of models, in which chiral ciliary flow provides the symmetry-breaking event, requires that multiple phyla use completely different mechanisms, be it using cilia that are all motile such as in the medaka [93]; cilia that are motile and sensory cilia that are immobile, such as in zebrafish or mouse [13]; or in the complete absence of the necessary ciliated nodal structure at all, such as in chick or pig [34,37,35,36]. We have discussed elsewhere the degree to which the various models of asymmetry match the available data [16,17]. Our data here are consistent with many prior studies [87,40,94,53,95] showing that asymmetry mechanisms function long prior to neurulation, and demonstrate that several cytoskeletal proteins randomize only when introduced extremely early.
It may be tempting to interpret some of the results via the role of cytoskeleton in cilia. However, the timing experiments with the $\alpha$-tubulin mutant (Fig. 2) and the myosin Flailer mutant (Fig. 6) demonstrate that their disruptive function occurs immediately post-fertilization. The clear effect that the $\alpha$-tubulin mutant has on a late, neural crest-derived phenotype such as craniofacial development, regardless of the timepoint of injection, whilst losing its effect on laterality after 30mpf (Fig. 2), clearly demonstrates its ability to target cellular chirality and not ciliary function. Even an hour later, provided with many hours of opportunity to affect ciliary processes, $\alpha$-tubulin-T56I, Mgrn1-G2A and Mgrn1-C314D are not effective at randomizing asymmetry, definitively ruling out ciliary mechanisms as an explanation for the randomizing action of these particular proteins.
As before (Lobikin et al., 2012), we targeted different blastomeres at the 4-cell stage. Dorsal left (DL) blastomere contributes to the gastrocoel roof plate (GRP), as the GRP forms from dorsal blastomere descendants, and only cilia on the left side of the GRP are required for LR-relevant ciliary flow [96]; thus, phenotypes induced by ventral right (VR) blastomere injections would be independent of any effects on ciliary flow. The original lineage analysis [96] was recently challenged with conflicting data showing that cells derived from the VR blastomere can contribute to the immotile sensory cilia [97]. Regardless of which of those studies is correct, it is still the case that reagents injected into DL blastomeres are expected to affect ciliary flow at the GRP far more effectively than those injected into the VR blastomere, which is not the case for some of the reagents we tested, demonstrating how effects on asymmetry can diverge from effects predicted by the cilia models.
The cytoskeleton can generate chiral information de novo. Self-organization of the actin cytoskeleton has been shown to generate cellular chirality [98] and chirality in single cells [18–21], plants [23], snails [24,25], nematodes [26–28], fruit flies [29–33] and frog [45]. We propose that given our data linking this body of work to vertebrate development, the most parsimonious model is that of a conserved, intracellular, early origin of the LR axis, followed by subsequent elaboration events [17,44].
We investigated, at the subcellular level, the changes induced in embryonic cytoskeletal dynamics by the mutant proteins that randomize organ situs (Fig. 8) The data demonstrate that LR-relevant cytoskeletal molecules alter the behavior of microtubule plus ends. In future work, quantitative biophysical models of the cytoskeleton will be analyzed to understand precisely what cell properties are affected by the observed changes in plus end dynamics. Likely candidates include cells' physical properties (stiffness), motility, shape, and intracellular cargo delivery. Any of these parameters could affect downstream asymmetric gene expression and organogenesis, and multiscale models of early developmental biophysics, physiology, and transcriptional control will be built to address these questions quantitatively.
Laterality's many paths: asymmetric gene expression vs. organ situs
Our data allowed direct comparison between the degree of randomization of organ positioning vs. that observed in the expression of left-sided transcripts. While the canonical Nodal, Lefty, and Pitx2 genes are thought to be determinants of left-sidedness, not merely markers, our results highlighted some interesting new aspects of the LR pathway. First was the observation that treatments that randomize asymmetric gene expression strongly can have much weaker effects on organ situs. This reveals that these 3 genes are not definitive readouts of an embryo's laterality, and strongly suggests that future studies must score organs as well, not only assay by in situ hybridization to Nodal and similar probes [97,99]. The ability of many embryos to normalize organ locations despite incorrect asymmetric gene expression reveals the existence of a kind of self-repair or fixing checkpoint, which we have previously reported during craniofacial morphogenesis in Xenopus [100], and which is known to exist in a number of species [101]). Future work will identify the molecular components responsible for recognizing incorrect expression of genes like Nodal and correcting subsequent events. The discovery of these mechanisms, and why they operate in some but not all embryos, will likely have important implications for the biomedicine of laterality-related birth defects.
The dissociation between organ situs and Nodal expression that we observe with Mgrn1 (Fig. 3) has also been observed in mouse [47]. This, together with the similar outcome we showed in the class I myosins identified in Drosophila (Fig. 7), present an interesting challenge to the assumption that expression of laterality markers such as Nodal correlate exactly with organ situs (Fig. 9A). Nodal is thought to be a major instructive element in LR patterning, serving not only as a marker but being able to re-specify laterality in gain- and loss-of-function experiments [102–105,56]. However, the fact that in the same batch of embryos, up to 40% of embryos may exhibit incorrect Nodal expression sidedness but less than 10% organ reversals with some proteins (Fig. 3A,B) while Xnr1, Lefty and Pitx2 laterality can be closely predictive of organ situ in others (Fig. 5), suggests that Nodal signaling is not necessarily a definitive determinant of final morphological outcome. This has been noted in other pathways in multiple model systems [48] and the absent Nodal expression in embryos that grow up to be normal (7% of control embryos show no expression of Xnr1, e.g. Fig. 2C) that we have observed previously (see Fig. 7 in [53]) underscores that Nodal is not a definitive marker of laterality; these data suggest that Nodal expression is not a sufficient readout of laterality outcome – for some perturbations, its predictive value is not high.
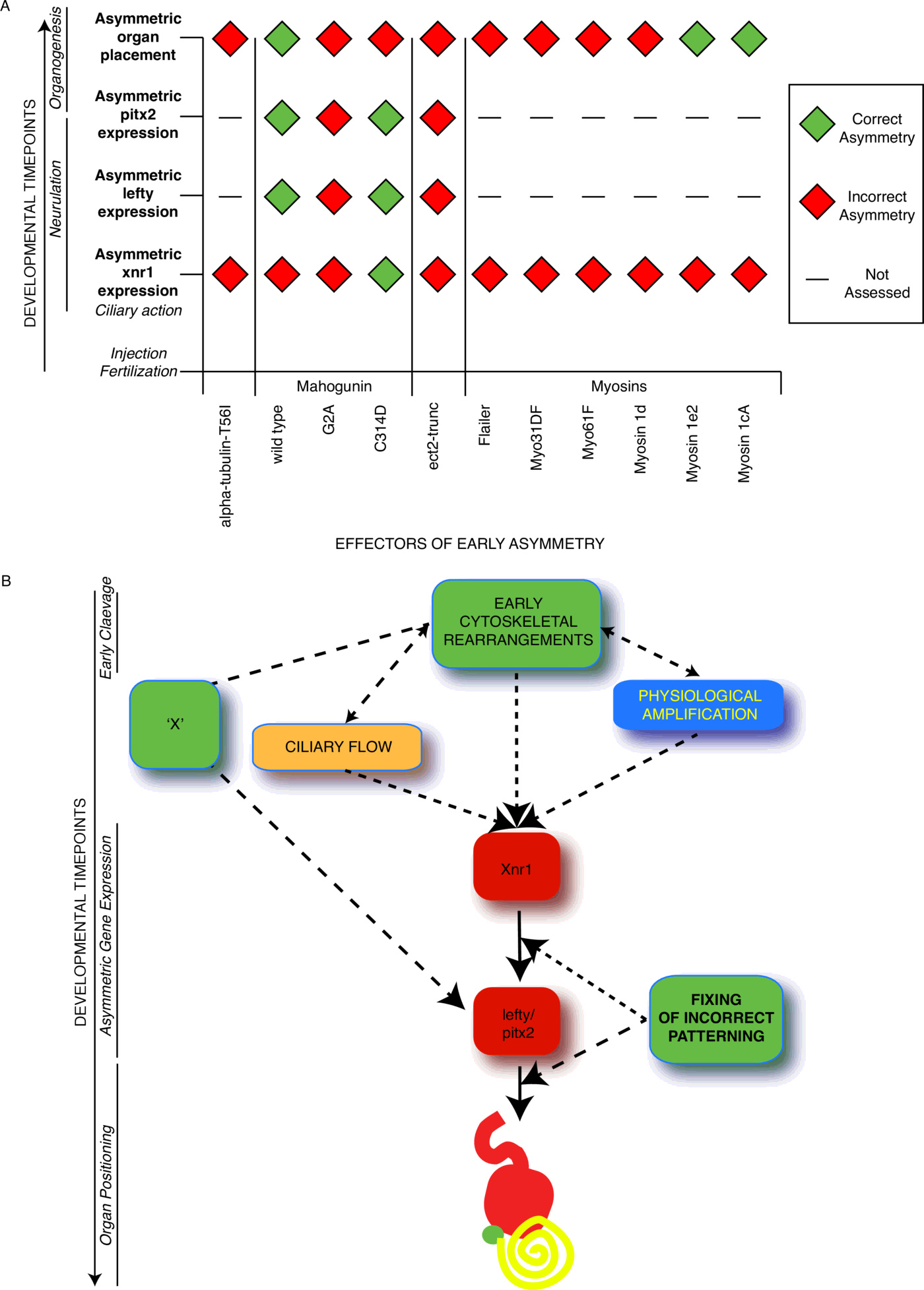
Conclusion
In Fig. 9, we summarize the discrepancies between Nodal asymmetry and organ asymmetry from our dataset and propose a model, whereby regulators of the early cytoskeletal role in LR patterning can not only affect Nodal positioning, but also organ patterning independent of the position of Nodal, via a parallel pathway. In this way, defects in LR patterning may be corrected during embryonic development, and this could explain the inability to induce the levels of heterotaxia expected if a discrete and single event is instructive in establishing this patterning. However, the mechanistic explanation for such discrepancies is not clear. Development of different organs at different timepoints of embryogenesis may require multiple levels of laterality signaling, from both early and late mechanisms.
It is clear that left-right asymmetry has only begun to reveal some of its true complexity. The error-correction, stochasticity, and redundancy observed in these data serve as powerful models for similar phenomena observed (but not yet explained) throughout development and biomedicine. LR patterning is a process that encompasses intracellular cytoskeletal events, bioelectric control of LR morphogens, ciliary sensing, planar cell polarity, organism-wide asymmetric transcription, and organ morphogenesis. It is likely that the eventual full unraveling of the control networks that integrates all of these steps will reveal fascinating new biology of wide relevance.
Acknowledgements
We thank Kenji Matsuno for Drosophila myosin (Myo31DF and Myo61F) cDNAs, Douglas Blackiston and other members of the Levin lab for useful discussions on these data, Dany Adams for discussions about craniofacial defects, Amanda Allen for laboratory assistance, and Erin Switzer for frog husbandry. L.A.L gratefully acknowledges support from NIH R00 MH095768. M.L. gratefully acknowledges funding support of the G. Harold and Leila Y. Mathers Charitable Foundation, and a subaward from the Physical Science Oncology Center supported by Award Number U54CA143876 from the National Cancer Institute.
Supplementary figures
- Supplementary Table 1: Organ situs data for mutant injections performed within 30mpf.
- Supplementary Table 2: Laterality of Xnr1 expression for mutant injections performed within 30mpf.
- Supplementary Table 3: Laterality of Lefty expression for mutant injections.
- Supplementary Table 4: Laterality of Pitx2 expression for mutant injections.
- Supplementary Table 5: Raw data from axon outgrowth plus-end microtubule tracking.
- Supplementary Table 6: Primers used in cloning
- Supplementary Figure 1: Comparison of organ heterotaxia and laterality of Xnr1 expression. (A) Organs incorrectly positioned in embryos injected within 30mpf. (B) Laterality of observed abnormal Xnr1 expression.
- Supplementary Figure 2: Examples of craniofacial defects observed for $\alpha$-tubulin-T56I-injected embryos
Peer-reviewed article version
A peer-reviewed version of this article can be found at Integrative Biology at the link:
http://pubs.rsc.org/en/content/articlelanding/2016/ib/c5ib00281h#!divAbstract
This is the preprint version that was submitted to the journal and does not contain revisions as a result of that peer-review process.
References
- J I Hoffman and S Kaplan. The incidence of congenital heart disease. J Am Coll Cardiol, 39(12):1890–1900, Jun 2002.
- M D Reller, M J Strickland, T Riehle-Colarusso, W T Mahle, and A Correa. Prevalence of congenital heart defects in metropolitan atlanta, 1998-2005. J Pediatr, 153(6):807–813, Dec 2008.
- J Burn. Disturbance of morphological laterality in humans. Ciba Found Symp, 162:282–96; discussion 296, Jan 1991.
- H Peeters and K Devriendt. Human laterality disorders. Eur J Med Genet, 49(5):349–362, Oct 2006.
- A F Ramsdell. Left-right asymmetry and congenital cardiac defects: getting to the heart of the matter in vertebrate left-right axis determination. Dev Biol, 288(1):1–20, Dec 2005.
- M Levin and A R Palmer. Left-right patterning from the inside out: widespread evidence for intracellular control. Bioessays, 29(3):271–287, Mar 2007.
- C Neville. Animal asymmetry. Edward Arnold, illustrated edition, 1976.
- A R Palmer. From symmetry to asymmetry: phylogenetic patterns of asymmetry variation in animals and their evolutionary significance. Proc Natl Acad Sci U S A, 93(25):14279–14286, Dec 1996.
- A R Palmer. Symmetry breaking and the evolution of development. Science, 306(5697):828–833, Oct 2004.
- C Tabin. Do we know anything about how left-right asymmetry is first established in the vertebrate embryo? J Mol Histol, 36(5):317–323, Jun 2005.
- L N Vandenberg and M Levin. Perspectives and open problems in the early phases of left-right patterning. Semin Cell Dev Biol, 20(4):456–463, Jun 2009.
- L N Vandenberg and M Levin. Far from solved: a perspective on what we know about early mechanisms of left-right asymmetry. Dev Dyn, 239(12):3131–3146, Dec 2010.
- B Basu and M Brueckner. Cilia multifunctional organelles at the center of vertebrate left-right asymmetry. Curr Top Dev Biol, 85:151–174, Jan 2008.
- T Okumura, H Utsuno, J Kuroda, E Gittenberger, T Asami, and K Matsuno. The development and evolution of left-right asymmetry in invertebrates: lessons from drosophila and snails. Dev Dyn, 237(12):3497–3515, Dec 2008.
- P Spéder, A Petzoldt, M Suzanne, and S Noselli. Strategies to establish left/right asymmetry in vertebrates and invertebrates. Curr Opin Genet Dev, 17(4):351–358, Aug 2007.
- L N Vandenberg, J M Lemire, and M Levin. It’s never too early to get it right: A conserved role for the cytoskeleton in left-right asymmetry. Commun Integr Biol, 6(6):e27155, Nov 2013.
- L N Vandenberg and M Levin. A unified model for left-right asymmetry? comparison and synthesis of molecular models of embryonic laterality. Dev Biol, 379(1):1–15, Jul 2013.
- T H Chen, J J Hsu, X Zhao, C Guo, M N Wong, Y Huang, Z Li, A Garfinkel, C M Ho, Y Tintut, and et al. Left-right symmetry breaking in tissue morphogenesis via cytoskeletal mechanics. Circ Res, 110(4):551–559, Feb 2012.
- A M Heacock and B W Agranoff. Clockwise growth of neurites from retinal explants. Science, 198(4312):64–66, Oct 1977.
- L Q Wan, K Ronaldson, M Park, G Taylor, Y Zhang, J M Gimble, and G Vunjak-Novakovic. Micropatterned mammalian cells exhibit phenotype-specific left-right asymmetry. Proc Natl Acad Sci U S A, 108(30):12295–12300, Jul 2011.
- J Xu, A Van Keymeulen, N M Wakida, P Carlton, M W Berns, and H R Bourne. Polarity reveals intrinsic cell chirality. Proc Natl Acad Sci U S A, 104(22):9296–9300, May 2007.
- T Hashimoto. Molecular genetic analysis of left-right handedness in plants. Philos Trans R Soc Lond, B, Biol Sci, 357(1422):799–808, Jun 2002.
- S Thitamadee, K Tuchihara, and T Hashimoto. Microtubule basis for left-handed helical growth in arabidopsis. Nature, 417(6885):193–196, May 2002.
- R Kuroda, B Endo, M Abe, and M Shimizu. Chiral blastomere arrangement dictates zygotic left-right asymmetry pathway in snails. Nature, 462(7274):790–794, Dec 2009.
- Y Shibazaki, M Shimizu, and R Kuroda. Body handedness is directed by genetically determined cytoskeletal dynamics in the early embryo. Curr Biol, 14(16):1462–1467, Aug 2004.
- E Frasnelli, G Vallortigara, and L J Rogers. Left-right asymmetries of behaviour and nervous system in invertebrates. Neurosci Biobehav Rev, 36(4):1273–1291, Apr 2012.
- S R Naganathan, S Fürthauer, M Nishikawa, F Jülicher, and S W Grill. Active torque generation by the actomyosin cell cortex drives left-right symmetry breaking. elife, 3:e04165, Dec 2014.
- C Pohl. Left-right patterning in the c. elegans embryo: Unique mechanisms and common principles. Commun Integr Biol, 4(1):34–40, Jan 2011.
- J B Coutelis, A G Petzoldt, P Spéder, M Suzanne, and S Noselli. Left-right asymmetry in drosophila. Semin Cell Dev Biol, 19(3):252–262, Jun 2008.
- C Géminard, N González-Morales, J B Coutelis, and S Noselli. The myosin id pathway and left-right asymmetry in drosophila. Genesis, 52(6):471–480, Jun 2014.
- J Kuroda, M Nakamura, M Yoshida, H Yamamoto, T Maeda, K Taniguchi, N Nakazawa, R Hatori, A Ishio, A Ozaki, and et al. Canonical wnt signaling in the visceral muscle is required for left-right asymmetric development of the drosophila midgut. Mech Dev, 128(11-12):625–639, Feb 2012.
- A G Petzoldt, J B Coutelis, C Géminard, P Spéder, M Suzanne, D Cerezo, and S Noselli. De-cadherin regulates unconventional myosin id and myosin ic in drosophila left-right asymmetry establishment. Development, 139(10):1874–1884, May 2012.
- K Taniguchi, R Maeda, T Ando, T Okumura, N Nakazawa, R Hatori, M Nakamura, S Hozumi, H Fujiwara, and K Matsuno. Chirality in planar cell shape contributes to left-right asymmetric epithelial morphogenesis. Science, 333(6040):339–341, Jul 2011.
- F Bangs, N Antonio, P Thongnuek, M Welten, M G Davey, J Briscoe, and C Tickle. Generation of mice with functional inactivation of talpid3, a gene first identified in chicken. Development, 138(15):3261–3272, Aug 2011.
- J Männer. Does an equivalent of the “ventral node” exist in chick embryos? a scanning electron microscopic study. Anat Embryol, 203(6):481–490, Jun 2001.
- Y Yin, F Bangs, I R Paton, A Prescott, J James, M G Davey, P Whitley, G Genikhovich, U Technau, D W Burt, and et al. The talpid3 gene (kiaa0586) encodes a centrosomal protein that is essential for primary cilia formation. Development, 136(4):655–664, Feb 2009.
- J Gros, K Feistel, C Viebahn, M Blum, and C J Tabin. Cell movements at hensen’s node establish left/right asymmetric gene expression in the chick. Science, 324(5929):941–944, May 2009.
- K Agata and Y Umesono. [evolution of the genetic program controlling brain development]. Tanpakushitsu Kakusan Koso, 44(3):245–249, Mar 1999.
- T Abe, S Thitamadee, and T Hashimoto. Microtubule defects and cell morphogenesis in the lefty1lefty2 tubulin mutant of arabidopsis thaliana. Plant Cell Physiol, 45(2):211–220, Feb 2004.
- M V Danilchik, E E Brown, and K Riegert. Intrinsic chiral properties of the xenopus egg cortex: an early indicator of left-right asymmetry? Development, 133(22):4517–4526, Nov 2006.
- M Nakamura and T Hashimoto. A mutation in the arabidopsis gamma-tubulin-containing complex causes helical growth and abnormal microtubule branching. J Cell Sci, 122(Pt 13):2208–2217, Jul 2009.
- S Schonegg, A Hyman, and W Wood. Timing and mechanism of the initial cue establishing handed left-right asymmetry incaenorhabditis elegans embryos. Genesis, 52(6):572–580, Jun 2014.
- P Spéder, G Adám, and S Noselli. Type id unconventional myosin controls left-right asymmetry in drosophila. Nature, 440(7085):803–807, Apr 2006.
- M Levin and N Nascone. Two molecular models of initial left-right asymmetry generation. Med Hypotheses, 49(5):429–435, Nov 1997.
- M Lobikin, G Wang, J Xu, Y W Hsieh, C F Chuang, J M Lemire, and M Levin. Early, nonciliary role for microtubule proteins in left-right patterning is conserved across kingdoms. Proc Natl Acad Sci U S A, 109(31):12586–12591, Jul 2012.
- T Okumura, T Sasamura, M Inatomi, S Hozumi, M Nakamura, R Hatori, K Taniguchi, N Nakazawa, E Suzuki, R Maeda, and et al. Class i myosins have overlapping and specialized functions in left-right asymmetric development in drosophila. Genetics, 199(4):1183–1199, Apr 2015.
- C D Cota, P Bagher, P Pelc, C O Smith, C R Bodner, and T M Gunn. Mice with mutations in mahogunin ring finger-1 (mgrn1) exhibit abnormal patterning of the left-right axis. Dev Dyn, 235(12):3438–3447, Dec 2006.
- L N Vandenberg. Laterality defects are influenced by timing of treatments and animal model. Differentiation, 83(1):26–37, Jan 2012.
- M Nakamura, K Matsumoto, Y Iwamoto, T Muguruma, N Nakazawa, R Hatori, K Taniguchi, R Maeda, and K Matsuno. Reduced cell number in the hindgut epithelium disrupts hindgut left-right asymmetry in a mutant of pebble, encoding a rhogef, in drosophila embryos. Mech Dev, 130(2-3):169–180, Feb 2013.
- J M Jones, J D Huang, V Mermall, B A Hamilton, M S Mooseker, A Escayg, N G Copeland, N A Jenkins, and M H Meisler. The mouse neurological mutant flailer expresses a novel hybrid gene derived by exon shuffling between gnb5 and myo5a. Hum Mol Genet, 9(5):821–828, Mar 2000.
- H L Sive, R M Grainger, and R M Harland. Early Development of Xenopus Laevis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, 2000.
- P D Nieuwkoop and J Faber. Normal Table of Xenopus Laevis. Garland Science, 1st edition, 1994.
- M Levin and M Mercola. Gap junctions are involved in the early generation of left-right asymmetry. Dev Biol, 203(1):90–105, Nov 1998.
- R M Harland. In situ hybridization: an improved whole-mount method for xenopus embryos. Methods Cell Biol, 36:685–695, Jan 1991.
- A H Monsoro-Burq. A rapid protocol for whole-mount in situ hybridization on xenopus embryos. CSH Protoc, 2007:pdb.prot4809, Aug 2007.
- K Sampath, A M Cheng, A Frisch, and C V Wright. Functional differences among xenopus nodal-related genes in left-right axis determination. Development, 124(17):3293–3302, Sep 1997.
- C Meno, Y Ito, Y Saijoh, Y Matsuda, K Tashiro, S Kuhara, and H Hamada. Two closely-related left-right asymmetrically expressed genes, lefty-1 and lefty-2: their distinct expression domains, chromosomal linkage and direct neuralizing activity in xenopus embryos. Genes Cells, 2(8):513–524, Aug 1997.
- M Campione, H Steinbeisser, A Schweickert, K Deissler, F van Bebber, L A Lowe, S Nowotschin, C Viebahn, P Haffter, M R Kuehn, and et al. The homeobox gene pitx2: mediator of asymmetric left-right signaling in vertebrate heart and gut looping. Development, 126(6):1225–1234, Mar 1999.
- L A Lowery, A E Faris, A Stout, and D Van Vactor. Neural explant cultures from xenopus laevis. J Vis Exp, (68):e4232, Oct 2012.
- K T Applegate, S Besson, A Matov, M H Bagonis, K Jaqaman, and G Danuser. plustiptracker: Quantitative image analysis software for the measurement of microtubule dynamics. J Struct Biol, 176(2):168–184, Nov 2011.
- L A Lowery, A Stout, A E Faris, L Ding, M A Baird, M W Davidson, G Danuser, and D Van Vactor. Growth cone-specific functions of xmap215 in restricting microtubule dynamics and promoting axonal outgrowth. Neural Dev, 8:22, Dec 2013.
- A Stout, S D’Amico, T Enzenbacher, P Ebbert, and L A Lowery. Using plustiptracker software to measure microtubule dynamics in xenopus laevis growth cones. J Vis Exp, (91):e52138, Sep 2014.
- T Ishida and T Hashimoto. An arabidopsis thaliana tubulin mutant with conditional root-skewing phenotype. J Plant Res, 120(5):635–640, Sep 2007.
- T Ishida, Y Kaneko, M Iwano, and T Hashimoto. Helical microtubule arrays in a collection of twisting tubulin mutants of arabidopsis thaliana. Proc Natl Acad Sci U S A, 104(20):8544–8549, May 2007.
- J L Lohr, M C Danos, and H J Yost. Left-right asymmetry of a nodal-related gene is regulated by dorsoanterior midline structures during xenopus development. Development, 124(8):1465–1472, Apr 1997.
- M R Rebagliati, R Toyama, C Fricke, P Haffter, and I B Dawid. Zebrafish nodal-related genes are implicated in axial patterning and establishing left-right asymmetry. Dev Biol, 199(2):261–272, Jul 1998.
- C Janke and J C Bulinski. Post-translational regulation of the microtubule cytoskeleton: mechanisms and functions. Nat Rev Mol Cell Biol, 12(12):773–786, Dec 2011.
- G S McDowell and A Philpott. Non-canonical ubiquitylation: Mechanisms and consequences. Int J Biochem Cell Biol, 45(8):1833–1842, Aug 2013.
- L Sun and Z J Chen. The novel functions of ubiquitination in signaling. Curr Opin Cell Biol, 16(2):119–126, Apr 2004.
- D Srivastava and O Chakrabarti. Mahogunin-mediated $\alpha$-tubulin ubiquitination via noncanonical k6 linkage regulates microtubule stability and mitotic spindle orientation. Cell Death Dis, 5:e1064, Feb 2014.
- A Upadhyay, A Amanullah, D Chhangani, R Mishra, A Prasad, and A Mishra. Mahogunin ring finger-1 (mgrn1), a multifaceted ubiquitin ligase: Recent unraveling of neurobiological mechanisms. Mol Neurobiol, Aug 2015.
- J Jiao, H Y Kim, R R Liu, C A Hogan, K Sun, L M Tam, and T M Gunn. Transgenic analysis of the physiological functions of mahogunin ring finger-1 isoforms. Genesis, 47(8):524–534, Aug 2009.
- D D Guerra, R Pratelli, E Kraft, J Callis, and G Pilot. Functional conservation between mammalian mgrn1 and plant log2 ubiquitin ligases. FEBS Lett, 587(21):3400–3405, Nov 2013.
- C Meno, Y Saijoh, H Fujii, M Ikeda, T Yokoyama, M Yokoyama, Y Toyoda, and H Hamada. Left-right asymmetric expression of the tgf beta-family member lefty in mouse embryos. Nature, 381(6578):151–155, May 1996.
- M Logan, S M Pagán-Westphal, D M Smith, L Paganessi, and C J Tabin. The transcription factor pitx2 mediates situs-specific morphogenesis in response to left-right asymmetric signals. Cell, 94(3):307–317, Aug 1998.
- J McGrath, S Somlo, S Makova, X Tian, and M Brueckner. Two populations of node monocilia initiate left-right asymmetry in the mouse. Cell, 114(1):61–73, Jul 2003.
- D M Supp, D P Witte, S S Potter, and M Brueckner. Mutation of an axonemal dynein affects left-right asymmetry in inversus viscerum mice. Nature, 389(6654):963–966, Oct 1997.
- A Armakolas and A J Klar. Left-right dynein motor implicated in selective chromatid segregation in mouse cells. Science, 315(5808):100–101, Jan 2007.
- A J Klar. Support for the selective chromatid segregation hypothesis advanced for the mechanism of left-right body axis development in mice. Breast Dis, 29:47–56, Jan 2008.
- Y Zhang and M Levin. Left-right asymmetry in the chick embryo requires core planar cell polarity protein vangl2. Genesis, 47(11):719–728, Nov 2009.
- A Wynshaw-Boris. Lissencephaly and lis1: insights into the molecular mechanisms of neuronal migration and development. Clin Genet, 72(4):296–304, Oct 2007.
- P Sitaram, M A Anderson, J N Jodoin, E Lee, and L A Lee. Regulation of dynein localization and centrosome positioning by lis-1 and asunder during drosophila spermatogenesis. Development, 139(16):2945–2954, Aug 2012.
- B Zimdahl, T Ito, A Blevins, J Bajaj, T Konuma, J Weeks, C S Koechlein, H Y Kwon, O Arami, D Rizzieri, and et al. Lis1 regulates asymmetric division in hematopoietic stem cells and in leukemia. Nat Genet, 46(3):245–252, Mar 2014.
- K D Sumigray, H Chen, and T Lechler. Lis1 is essential for cortical microtubule organization and desmosome stability in the epidermis. J Cell Biol, 194(4):631–642, Aug 2011.
- C W Sipe, L Liu, J Lee, C Grimsley-Myers, and X Lu. Lis1 mediates planar polarity of auditory hair cells through regulation of microtubule organization. Development, 140(8):1785–1795, Apr 2013.
- T Pramparo, O Libiger, S Jain, H Li, Y H Youn, S Hirotsune, N J Schork, and A Wynshaw-Boris. Global developmental gene expression and pathway analysis of normal brain development and mouse models of human neuronal migration defects. PLoS Genet, 7(3):e1001331, Mar 2011.
- T D Bunney, A H De Boer, and M Levin. Fusicoccin signaling reveals 14-3-3 protein function as a novel step in left-right patterning during amphibian embryogenesis. Development, 130(20):4847–4858, Oct 2003.
- A Kondratova, N Neznanov, R Kondratov, and A Gudkov. Poliovirus protein 3a binds and deregulates lis1, causing block of membrane protein trafficking and deregulation of cell division. Cell Cycle, 4(10):1403–1410, Oct 2005.
- S Prokopenko, R Saint, and H Bellen. Tissue distribution of pebble rna and pebble protein during drosophila embryonic development. Mech Dev, 90(2):269–273, Feb 2000.
- S Hozumi, R Maeda, K Taniguchi, M Kanai, S Shirakabe, T Sasamura, P Spéder, S Noselli, T Aigaki, R Murakami, and et al. An unconventional myosin in drosophila reverses the default handedness in visceral organs. Nature, 440(7085):798–802, Apr 2006.
- S M Schumacher-Bass, E D Vesely, L Zhang, K E Ryland, D P McEwen, P J Chan, C R Frasier, J C McIntyre, R M Shaw, and J R Martens. Role for myosin-v motor proteins in the selective delivery of kv channel isoforms to the membrane surface of cardiac myocytes. Circ Res, 114(6):982–992, Mar 2014.
- E A Bearce, B Erdogan, and L A Lowery. Tipsy tour guides: how microtubule plus-end tracking proteins (+tips) facilitate axon guidance. Front Cell Neurosci, 9:241, Jun 2015.
- K Kamura, D Kobayashi, Y Uehara, S Koshida, N Iijima, A Kudo, T Yokoyama, and H Takeda. Pkd1l1 complexes with pkd2 on motile cilia and functions to establish the left-right axis. Development, 138(6):1121–1129, Mar 2011.
- R L Gardner. Normal bias in the direction of fetal rotation depends on blastomere composition during early cleavage in the mouse. PLoS ONE, 5(3):e9610, Mar 2010.
- M Levin, T Thorlin, K R Robinson, T Nogi, and M Mercola. Asymmetries in h+/k+-atpase and cell membrane potentials comprise a very early step in left-right patterning. Cell, 111(1):77–89, Oct 2002.
- P Vick, A Schweickert, T Weber, M Eberhardt, S Mencl, D Shcherbakov, T Beyer, and M Blum. Flow on the right side of the gastrocoel roof plate is dispensable for symmetry breakage in the frog xenopus laevis. Dev Biol, 331(2):281–291, Jul 2009.
- M Tingler, T Ott, J Tözser, S Kurz, M Getwan, M Tisler, A Schweickert, and M Blum. Symmetry breakage in the frog xenopus: role of rab11 and the ventral-right blastomere. Genesis, 52(6):588–599, Jun 2014.
- Y H Tee, T Shemesh, V Thiagarajan, R F Hariadi, K L Anderson, C Page, N Volkmann, D Hanein, S Sivaramakrishnan, M M Kozlov, and et al. Cellular chirality arising from the self-organization of the actin cytoskeleton. Nat Cell Biol, 17(4):445–457, Apr 2015.
- A Schweickert, P Vick, M Getwan, T Weber, I Schneider, M Eberhardt, T Beyer, A Pachur, and M Blum. The nodal inhibitor coco is a critical target of leftward flow in xenopus. Curr Biol, 20(8):738–743, Apr 2010.
- L N Vandenberg, D S Adams, and M Levin. Normalized shape and location of perturbed craniofacial structures in the xenopus tadpole reveal an innate ability to achieve correct morphology. Dev Dyn, 241(5):863–878, May 2012.
- N Farinella-Ferruzza. The transformation of a tail into limb after xenoplastic transplantation. Experientia, 12(8):304–305, Aug 1956.
- K Fuller, J T O’Connell, J Gordon, O Mauti, and J Eggenschwiler. Rab23 regulates nodal signaling in vertebrate left-right patterning independently of the hedgehog pathway. Dev Biol, 391(2):182–195, Jul 2014.
- M Levin, R L Johnson, C D Stern, M Kuehn, and C Tabin. A molecular pathway determining left-right asymmetry in chick embryogenesis. Cell, 82(5):803–814, Sep 1995.
- T Nakamura, N Mine, E Nakaguchi, A Mochizuki, M Yamamoto, K Yashiro, C Meno, and H Hamada. Generation of robust left-right asymmetry in the mouse embryo requires a self-enhancement and lateral-inhibition system. Dev Cell, 11(4):495–504, Oct 2006.
- A Raya, Y Kawakami, C Rodriguez-Esteban, D Buscher, C M Koth, T Itoh, M Morita, R M Raya, I Dubova, J G Bessa, and et al. Notch activity induces nodal expression and mediates the establishment of left-right asymmetry in vertebrate embryos. Genes Dev, 17(10):1213–1218, May 2003.