From cytoskeletal dynamics to organ asymmetry: a non-linear, regulative pathway underlies left-right patterning
Version 1 Released on 09 May 2016 under Creative Commons Attribution 4.0 International LicenseAuthors' affiliations
- Department of Biology, School of Arts and Sciences - Tufts University
- Allen Discovery Center at Tufts University *. Unregistered author (unverified)
Keywords
Abstract
Consistent left-right asymmetry is a fundamental aspect of the bodyplan across phyla, and errors of laterality form an important class of human birth defects. Its molecular underpinning was first discovered as a sequential pathway of left- and right-sided gene expression that controlled positioning of the heart and visceral organs. Recent data have revised this picture in two important ways. First, the physical origin of chirality has been identified; cytoskeletal dynamics underlie the asymmetry of single cell behavior and of patterning of the left-right axis. Second, the pathway is not linear: early disruptions that alter the normal sidedness of upstream asymmetric genes do not necessarily induce defects in the laterality of the downstream genes or in organ situs. Thus, the LR pathway is a unique example of two fascinating aspects of biology: the interplay of physics and genetics in establishing large-scale anatomy, and regulative (shape-homeostatic) pathways that correct errors of patterning over time. Here, we review aspects of asymmetry from its intracellular, cytoplasmic origins to the recently-uncovered ability of the LR control circuitry to achieve correct gene expression and morphology despite reversals of key “determinant” genes. We provide novel functional data, in Xenopus laevis, on conserved elements of the cytoskeleton that drive asymmetry, and repair of downstream gene expression anomalies over developmental time. LR patterning can thus serve as a paradigm of how subcellular physics and gene expression cooperate to achieve developmental robustness of a body axis.
Introduction
Most vertebrates (and many invertebrates) have bilaterally-symmetric external bodyplans. Yet these same animals exhibit consistent asymmetries in the position or anatomy of internal organs such as heart, viscera, and brain [1]. Defects in LR asymmetry are an important class of human birth defects, including heterotaxia (the lack of concordance between internal organs, allowing each organ to individually ‘decide’ on its placement on the left or right side of the body), single organ inversions such as dextrocardia (the reversal in position and morphology of the heart), and isomerisms (symmetry of the LR axis, leading to either duplication or complete loss of unpaired organs such as the spleen). Patients with complete reversal of asymmetry (situs inversus) have fewer health consequences than these other conditions because heterotaxia and isomerisms often involve inappropriate connections between the heart, lungs and other visceral organs [2–6]. Interestingly, consistent laterality affects not only the asymmetric organs, but also the manifestation of numerous diseases and conditions affecting paired (seemingly symmetrical) organs, such as hip dysplasia, limb defects, and eye development [7–11]. This includes location and incidence of tumors [12–17], immune responses [18,19], and innervation of sensory organs such as eyes [20].
LR asymmetry is a unique puzzle in development [21], in complex organisms and even in colonies of simpler ones [22]. The embryonic anterior-posterior (AP) axis probably exists in order to place sense organs at the end of the animal that encounters novel environments first (oriented with the main axis of motion). The dorso-ventral (DV) axis can be set by gravity or sperm entry point. However, once those two orthogonal axes are set, the alignment of the LR axis is fixed; in order to distinguish L from R, symmetry has to be broken. Crucially, it is not merely a question of making L different from R, but doing it so that the LR axis is consistently oriented with respect to the AP and DV axes. The former process results in fluctuating asymmetry (an indicator of stress via the difficulty of keeping the two halves of the body precisely coordinated during growth); the latter is true biased asymmetry of anatomical structures. This is a difficult problem, as our universe does not macroscopically distinguish left from right [23,24]. This problem was noted long ago, by workers studying chiral biochemistry and its implications for development [25–27], clinical observations of mirror-imaging in anatomical features of human twins [28], unidirectional coiling of snail shells [29], and functional handedness in neural lateralization [30]. Since then, consistent LR asymmetry has been identified not only in anatomical structures from somites [31] to hair whorls [32–36] to deer antlers [37], but also in functional features such as immune response [38–40] and cancer [41,42,17,43]. Thus, the mechanisms of LR patterning are not only of fundamental interest in developmental biology, but are central to many interesting questions of physiology and evolution.
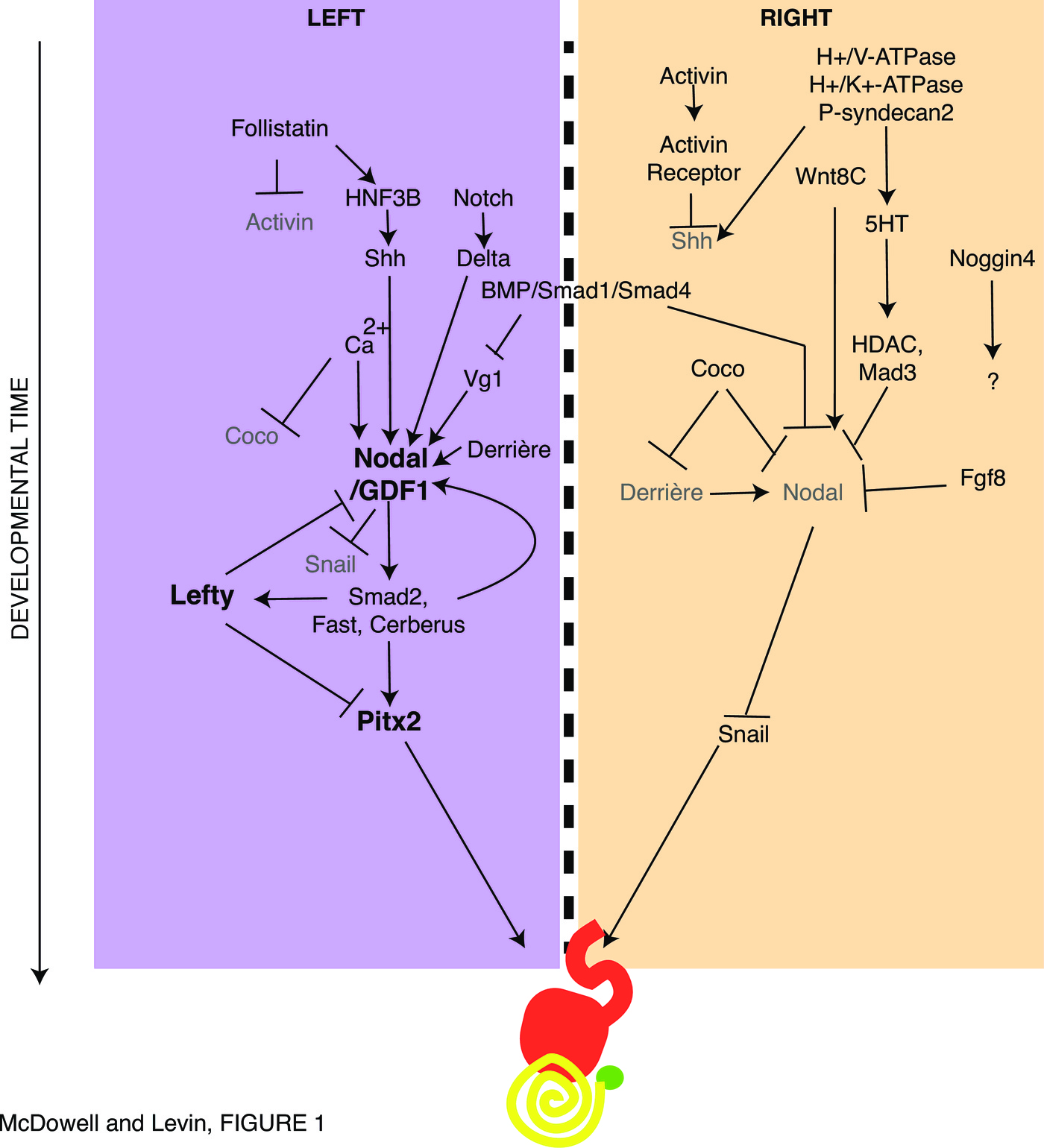
Early mechanistic work in this field identified a set of chemical agents that was able to perturb (randomize) asymmetry in animal models [44–48]. The first molecular explanations for the asymmetry of body organs came from studies in the chick [49], with the identification of asymmetrically-expressed genes, such as the left-sided Sonic hedgehog (Shh) and Nodal, the inductive and repressive relationships among these genes, and functional studies showing that aberrant expression of any of these was sufficient to randomize the situs of the heart, gut, and other viscera [50,51]. These data not only helped explain organ laterality in normal development but also provided a mechanistic explanation for laterality disturbances long known to occur in conjoined twins [52]. The central component of the LR pathway was the left-sided cassette formed by Shh inducing expression of Nodal inducing expression of Lefty inducing expression of Pitx2 [53–55].
The conserved molecular pathway in Xenopus, chick, mouse, and zebrafish as described in early work on LR asymmetry [56,57] has since been extended considerably, as described in Figure 1 and reviewed in [58,59], by loss- and gain-of-function approaches in a range of species that reveal the functional connections among LR patterning proteins. New players include Activin, Follistatin, Derrière, Coco, Mad3, BMP, Noggin4, and Fgf8 [60–65,53,66–70]. Recently, other downstream factors have been increasingly demonstrated in unilateral function during LR determination, such as calcium signaling and retinoic acid [71–79]. Together, this body of work reveals a progressive cascade of left- and right-specific activities that involve powerful signaling pathways. These signaling molecules then provide distinct signals to organ primordia on either side of the midline, resulting in asymmetric organogenesis.
Physics upstream and downstream of transcriptional networks
Regardless of the completeness of a transcriptional pathway or network model, one has to explain why the first asymmetric gene becomes expressed on one side but not the other. It is a fundamental limitation of transcriptional regulatory networks that they do not in themselves constrain geometry — no gene regulatory network (GRN) can generate consistently chiral output (distinguish left from right). Thus, upstream of any such regulatory cascade must lie some piece of physics which is able to break symmetry and reliably orient subsequent events with respect to the other two axes [80,81].
Interestingly, this is not strictly a multicellular phenomenon, in a number of systems, from bacteria to ciliates to human somatic cells in vitro, single cells exhibit consistent chirality in their movements and behaviors [25,82–95], both as individual cells and as collectives [92,96]. For example, outgrowth of neurites is predominantly clockwise [84,85,97,90]. The asymmetric growth and migration of various cell types on micropatterned surfaces has demonstrated the role of actomyosin networks in the generation and maintenance of consistent chirality of migration, cell shape, and tissue morphogenesis [82,91]. Thus, metazoan organisms face the problem of amplifying subcellular chirality into tissue-wide asymmetry across a midline, but do not need to reinvent the symmetry-breaking and orientation steps, as these appear to be ancient and ubiquitous.
One candidate for a chiral element of physics upstream of asymmetric gene expression is ciliary flow at neurulation [72,98,99]; this model however faces numerous problems which have been detailed elsewhere [100–104]. Recent functional data revealed conserved mechanisms in a range of organisms from plants to mammals, that establish asymmetry without a ciliated structure, or long before it forms; indeed, many phyla including some vertebrates determine their LR axis very early after fertilization [105–115]. Even mouse embryos are known to exhibit molecular and functional asymmetries (e.g., components such as cofilin, which is also asymmetric in cleavage-stage frog embryos) as early as the cleavage stages [107,116,117].
While ciliary flow may impinge on downstream transcriptional events in those species where it exists (not pig [108] or chick [118,119] for example), it cannot be the origin of asymmetry in most phyla. From data in a range of model species, it is clear that numerous aspects of development, including maternal protein localization [120,121,111,122], Wnt signaling [123], and small signaling molecules [114] are already consistently asymmetric long before cilia appear: most animal embryos can tell their Left from their Right at very early stages. Thus, the search for the origin of asymmetry has been extended far upstream of neurulation [124,47].
One interesting class of models links asymmetry to chromatid segregation [125–128] – a proposal that needs to be tested in the available model systems, as it offers the possibility of linking asymmetry to the fundamental dynamics of DNA. This model has been linked to data on birth defects and epithelial morphogenesis in humans and mice [129]. As in Xenopus, the mouse model has also revealed that $\alpha$-tubulin organization is critical for asymmetry, via studies of the protein Mahogunin [130,105,111,112,131].
A remarkably prescient prediction was made by a paper published before all of the molecular work on the LR pathway [132], in which Brown and Wolpert hypothesized a chiral element that initiates biased transport inside the early embryonic blastomeres. More recent work in several models have confirmed early hypotheses while elaborating on these ideas [133,134], showing that the cytoskeleton is centrally involved in generating the original asymmetric cues that break and orient symmetry. A number of model species such as C. elegans [135,74,136–138] and snails [139–142] were known to use cytoskeletal dynamics to determine chiral cell behavior and subsequent LR patterning. However recent data in these models, together with mammalian cells and Drosophila [143,144], have revealed many of the key details.
The intriguing structure of centrioles [145], including their anti-clockwise rotation [146], and the role of microtubules in generating asymmetry in neutrophils [147,93], plants [148], and frog [111] and C. elegans embryos [149,150], lends evidence to the theory that the centrosome could be a symmetry-breaking, chiral structure [100,151]. Evidence for intrinsic chirality, and not interactions with the substrate, were provided by the counter-clockwise rotation exhibited in zebrafish melanophores [94]. A particularly illustrative case of spontaneous intracellular chiral cytoskeletal organization again illustrates the role of actomyosin networks in the ability of $\alpha$-actinin to spontaneously arrange directionally, or reverse the directionality if the protein is grossly overexpressed [89].
Studies in Drosophila have demonstrated the effect of intrinsic cellular chirality on embryonic laterality [143,152–160]. Unconventional myosins, such as Myosin1d, have a clear role in affecting the asymmetry of the gut and genitalia in Drosophila [158,159] and this asymmetry is due to a direct effect on the actin filaments in epithelia [154]. Actin motility on Myo1c occurs in counterclockwise direction [161]; myosin V is a left-handed spiral motor toward the plus end of actin [162], while myosin II is right-handed spiral motor [163]. Moreover, the effect of these unconventional myosins on organismal asymmetry is linked to their effect on intrinsic cellular chirality [160], and individual cells can contribute to mechanical differences in generating chirality at the tissue level [153,160], a finding also demonstrated in Caenorhabditis elegans [113,134].
Our own work in Xenopus has demonstrated a number of key features that point to the importance of the cytoskeleton at multiple points in laterality. Firstly, fundamental cytoskeletal proteins such as tubulins and myosins are functionally important for normal embryonic laterality at the very earliest points of embryonic development, immediately post-fertilization [111,112]. The cytoskeleton appears to be required for the normal early localization of asymmetric components such as ion channels [121,111,122], a mechanism widely conserved to many somatic cell types [164,165]. Secondly, the conservation of phenotypes obtained from mutations in cytoskeletal and cytoskeleton-associated proteins exhibited in frog, compared to those of plants [111,112], snails [105], and Drosophila [112], demonstrate the need for physical signals upstream of gene regulatory networks (GRNs): for example, the organ-specific effects of myosins on Drosophila laterality were replicated in frog [112], transcending the differences in molecular LR pathways between vertebrates and invertebrates and raising the question of how the cytoskeleton generates chirality. Finally, the apparent disconnect between the asymmetric expression of Nodal and subsequent organ situs observed in mouse [130] and replicated in frog [112] highlight a conserved role for the cytoskeleton not only in laterality, but in the ability to correct earlier defects in laterality between the point of expression of markers of laterality and the positioning of visceral organs.
Downstream of the chiral physics within cells lie physiological mechanisms that amplify subcellular chiralities into true LR asymmetries across cell fields. One such system is the diffusion-based LALI (local activation, long range inhibition) system described in mouse [166,167]. Another is the chiral bioelectric gradient that redistributes intracellular morphogens such as maternal serotonin [120,168–171,110,172,78,173]. This process is most well-understood in frog embryos, but has also been observed and functionally implicated in amphioxus [174], sea urchin [168,175,176], C. elegans [177–179], zebrafish [120,180,181], and recently humans [182,183]. Indeed very recent analysis of the differences between blastomeres in the very earliest stages of embryo development, identified distinct metabolites in L vs. R cells in the 8-cell embryo using mass spectrometry [114]. The majority of these metabolites themselves have roles in functional regulation of ion transport [184–192], suggesting possible feedback loops in electrophysiology that could be important amplifying mechanisms for initially subtle LR asymmetry. Both of these systems impinge upon a key asymmetric gene – NODAL [193,62], and lie upstream of a cascade of asymmetric gene interactions. However, much as it needs a physical process to anchor consistent asymmetry upstream, genetic pathways likewise need to control physical forces in order to actually implement asymmetric morphogenesis.
GRNs feed into specific proteins that harness physical forces such as tension and adhesion to control asymmetric bending and growth of internal organs [194]. Mechanical forces are critical for the rotation and looping of internal organs ([195,196], see reviews for the heart in chick [197–200]). In the development of Xenopus gut morphology, a rightward torsion results in concave and convex topologies for cells on the left and right sides of the lateral plate mesoderm respectively, and cells on the right elongate twice as much as cells on the left, although proliferation rates remain the same [201]. Similarly, in Drosophila, asymmetries arising from planar cell polarity in gut looping through the activity of myosin1d were able to generate differences in tension, with greater tension on the left side of the cell driving leftward rotation [160]. In heart looping, a similar role for actin and myosin in driving dextral looping has been exhibited in zebrafish [196] and chick [202]. Recently computational modeling has supported evidence suggesting that differential growth supplies the forces that cause the heart tube to bend ventrally, while cytoskeletal movements can drive rightward torsion [203]. The maintenance of symmetry could also be due to differences in the composition of the extracellular matrix (ECM) as observed in perturbations of ECM composition in frog and chick embryos [204,48].
Developmental regeneration: robustness of the LR pathway
Bracketed upstream and downstream by interaction with physical forces, the middle of the LR patterning process consists of a pathway made up of sequentially interacting left- and right-sided gene products (Fig. 1). The implication of this kind of induction/repression model is that if something goes wrong upstream — if a particular gene is expressed on the wrong side for example — then the downstream elements will likewise be incorrect. For example, in the early work on LR determining genes in chick, it was shown that if the left-sided Shh gene was misexpressed on the right side, right-sided expression of the normally left-sided gene Nodal followed, and organs were subsequently randomized.
Importantly however, development in many species is highly regulative: early errors (e.g., splitting embryos in half [205]) can become subsequently corrected. While many perturbations overcome this basic shape homeostasis property (resulting in birth defects), it is nevertheless true that embryos are highly robust because of the ability to remodel after some kinds of deviations from the normal sequence of events. One example is the derivation of largely normal frog faces from early embryos with severely malformed facial structures, which self-correct over time [206]. The mechanisms of robustness and shape homeostasis are poorly understood at the mechanistic level, although they are now increasing objects of interest in molecular developmental biology [207–209] and regenerative medicine [210–212].
Self-correction capabilities in the LR pathway are a novel, molecularly-tractable example of regenerative repair. It is a uniquely-accessible context in which to study the regulatory mechanisms that recognize and reverse abnormal patterning states. Here, we identify new aspects of the cytoskeletal machinery that lie upstream of the LR asymmetry patterning pathway and investigate their robustness. We quantitatively analyze published data as well as our new functional studies to reveal remarkable flexibility in the GRN that belies a simple linear pathway to reveal remarkable pattern robustness.
Results: endogenous repairing of induced left-right defects
The canonical description of the left-right patterning pathway in vertebrates, regardless of the proposed point of symmetry breaking, follows the linear track of leftward activation of Nodal, downstream activation of Lefty and Pitx2, and thence to the correct placement of visceral organs. However, we have observed in our recent studies of the determination of laterality in Xenopus, many anomalies and exceptions to this neat pipeline.
For example, in studying proteins with roles in early embryonic asymmetries [213], and in particular cytoskeletal proteins with a conserved role in determining embryonic laterality [105,112], the number of embryos with reversed organs is actually smaller than the number with incorrect Nodal expression (Table 1). This suggests that the pathway from Nodal to organ situs is not as linear as had been assumed: having incorrect Nodal expression does not actually mean that an embryo’s organs will be reversed, despite Nodal’s established role as a determinant or “master regulator” of LR situs.
| Incorrect | ||||
| Protein | Nodal expression | Reversed Organs | Reference | Degree of repair |
| $\alpha$-tubulin T56I | 37% | 21% | [112] | 43% |
| (R: 10%, Bi: 9%, N: 18%) | ||||
| Lis-N99 | 26% | 18% | [112] | 30% |
| (R: 9%, Bi: 8%, N: 9%) | ||||
| Lis-C137 | 21% | 10% | [112] | 52% |
| (R: 7%, Bi: 9%, N: 5%) | ||||
| 14-3-3E | 65% | 30% | [112] | 54% |
| (R: 12%, Bi: 30%, N: 23%) |
| Incorrect | ||||
| Protein | Nodal expression | Reversed Organs | Reference | Degree of repair |
| Flailer | 57% | 18% | [112] | 68% |
| (R: 10%, Bi: 17%, N: 30%) | ||||
| Myo31DF | 24% | 12% | [112] | 50% |
| (R: 1%, Bi: 19%, N: 4%) | ||||
| Myo61F | 35% | 14% | [112] | 60% |
| (R: 5%, Bi: 16%, N: 14%) | ||||
| Myosin1d | 43% | 15% | [112] | 65% |
| (R: 9%, Bi: 8%, N: 26%) | ||||
| Myosin1e2 | 28% | 4% | [112] | 86% |
| (R: 7%, Bi: 4%, N: 17%) | ||||
| Myosin1cA | 44% | 2% | [112] | 96% |
| (R: 26%, Bi: 13%, N: 5%) |
| Incorrect Nodal expression | Reversed Organs | Reference | Degree of repair |
| 12% | 3-8% | [214] | 33-75% |
| spontaneous cardiac reversals | |||
| 34% | $<$1% | [186] | 97% |
| 7% | 1% | [112] | 86% |
| 0.5% | [105] | N/A |
Reviewing the literature for evidence of these phenomena, we found some interesting supportive observations. To begin with, there were a number of observations showing various degrees of correction or “fixing” of laterality defects from early to late markers (Table 4).
| Incorrect | ||||
| Protein | Nodal expression | Reversed Organs | Reference | Degree of repair |
| Cx26 2 of 4-cell: | 48% | 32% | [186] | 33% |
| ventral | (R: 1%, Bi: 1%, N: 46%) | |||
| Cx43 2 of 4-cell: | 32% | 22% | [186] | 31% |
| ventral | (R: 0%, Bi: 5%, N: 27%) | |||
| H7 2 of 4-cell: | 57% | 29% | [186] | 49% |
| ventral | (R: 5%, Bi: 14%, N: 38%) |
| Incorrect | Incorrect | Incorrect | Degree | |||
| Drug treatment | Nodal | Lefty | Pitx2 | Reversed | Reference | of |
| expression | expression | expression | Organs | repair | ||
| Lansoprazole | 36% | 40% | 47% | 51% | [110] | -42% |
| (R: 18%, | (R: 4%, | (R: 23%, | ||||
| Bi: 7%, | Bi: 32%, | Bi: 18%, | ||||
| N: 11%) | N: 4%) | N: 6%) | ||||
| HMR-1098 | 33% | 40% | [216] | -21% | ||
| (R: 4%, | ||||||
| Bi: 28%, | ||||||
| Concanamycin | 54% | 33% | [120] | 39% | ||
| (R: 8%, | ||||||
| Bi: 19%, | ||||||
| N: 27%) | ||||||
| Tropisetron | 36% | 16% | 23% | [170] | 36% | |
| (R: 6%, | (R: 2%, | |||||
| Bi: 15%, | Bi: 5%, | |||||
| N: 15%) | N: 9%) | |||||
| Anandamide | 53% | 35% | [186] | 34% | ||
| (R: 1%, | ||||||
| Bi: 3%, | ||||||
| N: 49%) | ||||||
| Heptanol | 81% | 48% | [186] | 41% | ||
| (R: 0%, | ||||||
| Bi: 0%, | ||||||
| N: 81%) | ||||||
| Glycyrrhetinic | 90% | 40% | [186] | 55% | ||
| acid | (R: 2%, | |||||
| Bi: 2%, | ||||||
| N: 86%) |
| Incorrect | ||||
| Protein | Nodal expression | Reversed Organs | Reference | Degree of repair |
| BVg1 R3 16-cell | 88% | 89% | [217] | -1% |
| tAR L1 16-cell | “complex” | 51% | [217] | Unknown |
| BMP2 L2 16-cell | 30% | 85/58% | [217] | Unclear |
| BMP2 R2 16-cell | 17.5% | 0% | [217] | Complete |
| correction | ||||
| Xwnt-8 L4 | 17% | 63% | [217] | -271% |
| Xwnt-8 L4, BVg1 R1 | 66% | 88% | [217] | -33% |
| Xwnt-8 L4, BVg L1 | “not determined” | 14% | [217] | Unknown |
| BVg1 L4 16-cell | 15% | 10% | [218] | 33% |
| BVg1 R4 16-cell | 75% | 52% | [218] | 31% |
| cardiac reversal | ||||
| Activin A PROTEIN | ||||
| Right-sided Injection | 83% | 49% | [219] | 41% |
| at Stage 20 | ||||
| Activin A PROTEIN | ||||
| Left-sided Injection | Not shown | 19% | [219] | Unknown |
| at Stage 20 |
| Incorrect | Incorrect | Degree | |||
| Drug treatment | Nodal | Lefty | Reversed | Reference | of |
| expression | expression | Organs | repair | ||
| GR113808 | 43% | 14% | 44% | [170] | -2% |
| (R: 10%, | (R: 3%, | ||||
| Bi: 26%, | Bi: 2%, | ||||
| N: 7%) | N: 9%) | ||||
| Iproniazid | 43% | 15% | 30% | [170] | 30% |
| (R: 10%, | (R: 2%, | ||||
| Bi: 21%, | Bi: 4%, | ||||
| N: 13%) | N: 10%) |
| Incorrect | Incorrect | Incorrect | Degree | ||
| Drug treatment | Nodal | Lefty | Pitx2 | Reversed | of |
| expression | expression | expression | Organs | repair | |
| Control | 7% | 3% | 4% | 1% | 86% |
| (R: 1%, | (R: 0%, | (R: 0%, | |||
| Bi: 2%, | Bi: 1%, | Bi: 2%, | |||
| N: 4%) | N: 2%) | N: 2%) | |||
| Mgrn wt | 44% | 8% | 4% | 6% | 91% |
| (R: 10%, | (R: 0%, | (R: 0%, | |||
| Bi: 23%, | Bi: 0%, | Bi: 0%, | |||
| N: 11%) | N: 8%) | N: 4%) | |||
| Mgrn G2A | 26% | 20% | 23% | 15% | 42% |
| (R: 4%, | (R: 3%, | (R: 6%, | |||
| Bi: 10%, | Bi: 1%, | Bi: 10%, | |||
| N: 12%) | N: 16%) | N: 7%) | |||
| Mgrn C314D | 14% | 8% | 14% | 17% | -21% |
| (R: 1%, | (R: 4%, | (R: 0%, | |||
| Bi: 8%, | Bi: 4%, | Bi: 7%, | |||
| N: 5%) | N: 0%) | N: 7%) | |||
| Ect2-trunc | 24% | 27% | 34% | 25% | -4% |
| (R: 6%, | (R: 10%, | (R: 12%, | |||
| Bi: 8%, | Bi: 2%, | Bi: 12%, | |||
| N: 10%) | N: 15%) | N: 10%) |
| Incorrect | Incorrect | Incorrect | Degree | ||
| Drug treatment | Nodal | Lefty | Pitx2 | Reversed | of |
| expression | expression | expression | Organs | repair | |
| Control | 8% | 14% | 0% | 1% | 88% |
| (R: 1%, | (R: 1%, | (R: 0%, | |||
| Bi: 2%, | Bi: 0%, | Bi: 0%, | |||
| N: 5%) | N: 13%) | N: 0%) | |||
| Mgrn wt | 37% | 7% | 9% | 3% | 92% |
| (R: 6%, | (R: 0%, | (R: 3%, | |||
| Bi: 23%, | Bi: 5%, | Bi: 3%, | |||
| N: 8%) | N: 2%) | N: 3%) | |||
| Mgrn C314D | 25% | 22% | 39% | 20% | 20% |
| (R: 6%, | (R: 0%, | (R: 5%, | |||
| Bi: 9%, | Bi: 12%, | Bi: 5%, | |||
| N: 10%) | N: 10%) | N: 29%) | |||
| Ect2-trunc | 44% | 50% | 48% | 28% | 36% |
| (R: 8%, | (R: 25%, | (R: 17%, | |||
| Bi: 17%, | Bi: 15%, | Bi: 9%, | |||
| N: 19%) | N: 10%) | N: 22%) |
Overexpression of Nodal and effects on laterality
The data above shows that when very early LR patterning steps are perturbed, and Nodal expression is incorrect, there can be a subsequent correction of this information downstream to lead to a lower number of tadpoles with reversed organs. However, we wanted to test whether direct misexpression of Nodal led to a clear mispatterning of laterality markers, and a reversal of organ situs, or whether misexpression of Nodal could also be corrected or tolerated by the developing embryo. What happens if Nodal is directly randomized, by forced misexpression of Nodal protein on the right side, or by overexpression of Nodal in general? A number of studies have already looked at the effect of Nodal overexpression in Xenopus, using both the injection of plasmid DNA [220,221] and mRNA [222] (summarized in Table 10).
| Incorrect | |||
| Treatment | Pitx2 expression | Reversed Organs | Reference |
| Xnr1 plasmids into 1 of 4 - left | 9% | [220] | |
| Xnr1 plasmids into 1 of 4 - right | 86% | [220] | |
| 50 pg Xnr1 into LPM dorsal blastomeres, | 85% | [222] | |
| 8-cell, bilateral | |||
| DR injections xnr1 20pg pXEX | 61% | [221] | |
| DR injections xnr1 100pg pXEX | 71% | [221] | |
| DR injections xnr1 20pg pCSKAX | 76% | [221] | |
| DR injections xnr1 100pg pCSKA | 68% | [221] | |
| DL injections xnr1 20pg pXEX | 20% | [221] | |
| DL injections xnr1 100pg pXEX | 25% | [221] | |
| DL injections xnr1 20pg pCSKA | 33% | [221] | |
| DL injections xnr1 100pgpCSKA | 50% | [221] |
| Incorrect | Incorrect | ||
| Lefty expression | Pitx2 expression | Reversed Organs | Degree of Repair |
| 61% | 61% | 39% | 36% |
| (R: 27%, Bi: 32%, N: 2%) | (R: 20%, Bi: 40%, N: 1%) |
| Organ placement | Left-sided | Right-sided | Both sides |
| Situs solitus | 158 | 70 | 28 |
| Heterotaxia | 22 | 100 | 34 |
Discussion
Gene-regulatory cascades form an important part of the middle phases of LR patterning [102], but important elements of physics and physiology lie upstream, in determining the sidedness of the first asymmetric transcription, and downstream, in implementing laterality cues toward asymmetric morpogenesis. Early events feed into different parts of the LR cascade, such as overexpression of Wnt8 and of the C314D mutant of mahogunin which is unable to ubiquitylate -tubulin. Indeed, some aspects of tissue morphogenesis may be intrinsic, using cell-level chirality to implement asymmetric looping directly, as may occur in Drosophila [96] and zebrafish [196].
While many organisms establish large-scale asymmetries, it is interesting that chirality is fundamental to individual cells – an ancient evolutionary feature that metazoans exploit for macroscopic anatomical purposes. Aside from highlighting the propagation of properties across orders of magnitude of scale, laterality sheds light on the relationship between genome and fundamentally epigenetic factors. In single-cell ciliates, Beisson and Sonneborn demonstrated that reversal of a ciliary row in the cell cortex is propagated to offspring. Their function is also reversed, and cells eventually starve because food particles are swept into the wrong direction; their normal genome is powerless to rescue them [223–226].
The original picture of the LR cascade made use of a midline barrier which separated distinct Left- and Right-sided programs of repression and induction [227]. This was subsequently revised by the finding that the L and R sides needed to communicate long-range via gap junction-mediated physiological signals for proper expression of early asymmetric genes [186,228], but the importance of establishing a robust midline was clear [229–233]; indeed, the LR axis cannot be oriented without a midline that sets the axis of symmetry. While some animals are thought to establish the midline later than others, and the LR axis is often viewed as being defined after and with respect to the AP and DV axes, cleavage patterns and the prevalence of strictly bilateral gynandropmorphs resulting from very early cleavage events reveal that from insects to man [234–236], separating the L and R sides is one of the first things most embryos do [237]. Once the midline separates L and R compartments and they establish their unique identities, the standard model holds that cascades of genes become expressed sequentially, in a functional pathway that should propagate errors as genes inappropriately expressed on one side exert their effects and turn on/off downstream genes counter to their normal restricted unilateral patterns.
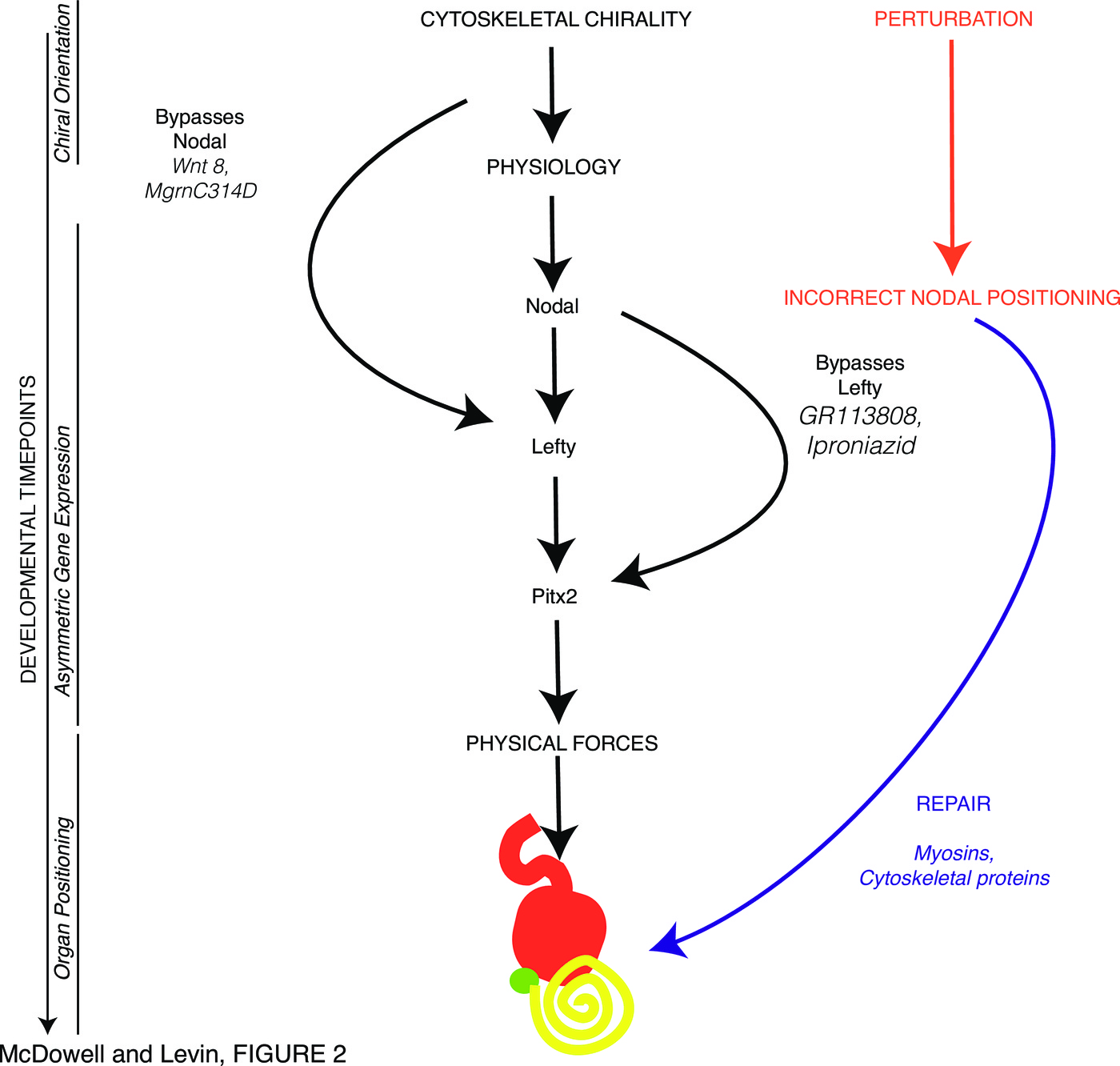
Most importantly, we have found, in both our new data and data from published experiments in Xenopus, discrepancies between the incidence of incorrect expression of early laterality markers and that of abnormal positioning of organs; these examples reveal the ability of embryos to correct defects in LR patterning over time (Figure 2). For example, early misexpression of unconventional myosin proteins (Table 2) and the mahogunin protein which targets $\alpha$-tubulin for degradation (Tables 8,9) strongly disrupt the normal laterality of Nodal expression but have no effect on the positioning of organs in those embryos.
In the case of low-frequency vibrations that induce LR patterning defects in early embryos, it appears that the early (cleavage) stages are crucial for enabling downstream repair mechanisms post-Nodal. Vandenberg et al. found that the level of Nodal expression randomization was constant and high despite the stage at which treatment began (as long as treatment occurred prior to blastula stage), but in all cases the percentage of resulting tadpoles with mispatterned organs was lower ([238], summarized in Figure 3A). This underscores the potential divergence between organ endpoints and marker expression in experiments with different timing conditions, and suggests that future studies cannot simply use gene expression readouts but must score organs as well to get a true picture of the effects of specific perturbations on the LR pathway. These data suggest that the earliest events (from 1-cell to st. 6) are important to enabling whatever mechanisms allow organs to form correctly despite abnormal Nodal laterality, as the ability to repair organ positioning increases progressively with treatments that start at later cleavage stages.
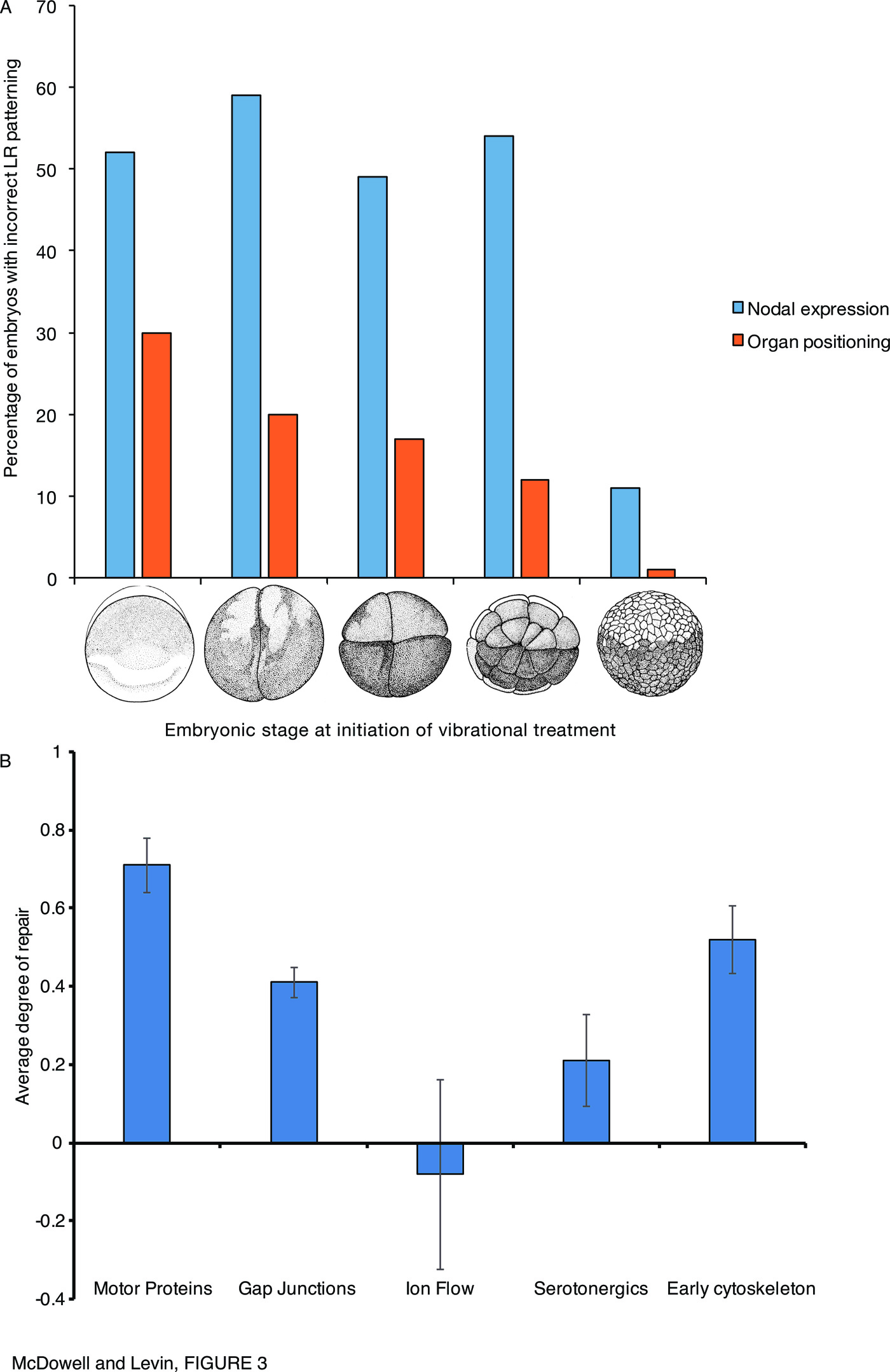
Interestingly, grouping the experimental results on normalization of downstream steps by the type of early perturbation (Figure 3B, Supplementary Table 1) suggests that the degree of laterality repair capability varies depending on the type of perturbation (the LR pathway target that was perturbed to generate the Nodal randomization). Perturbations of motor proteins, and the early cytoskeleton, within the first cell division, is tolerable to a certain degree as it largely becomes normalized by the time of organogenesis; but manipulations of ion flows are apparently not possible to recover from. It should also be noted that in most cases within the serotonergic group, Lefty appeared to be bypassed (Table 7). These data suggest that further study into the different degree of pathway repair downstream of targeting different types of components of LR patterning may shed greater light on the mechanism, and robustness, of LR patterning and the normalization of downstream targets despite randomized prior steps.
The data also reveal non-linearity of the pathway as some elements can be bypassed: interfering with early serotonin and monoamine oxidase signaling results in organ-level LR defects despite the correct expression of Lefty (Table 7). A dynamical systems view of these repair pathways, as a network rather than a linear pathway with “necessary and sufficient” master regulators, is necessary, as has already been noticed in the field of cancer and the search for driver genes [241–246]. The future is likely to involve not only molecular-genetic picture of these repair pathways, but a cybernetic, systems-control view of the information processed by these closed-loop, shape-homeostatic capabilities of embryogenesis [247–251]. It thus becomes clear that checking immediate downstream consequences of gene misexpression is not sufficient for functional analyses; a subtler strategy targeting further upstream of a gene of interest, which gives the embryo more time to recognize errors, is required to form a full picture of regulatory networks and “necessary and sufficient” claims for an explanation of what gene product determines expression of some other gene product.
In a sense, pattern homeostasis, such as seen in reparative regeneration widely across the tree of life [210], could be seen as a primary biological capability. Development is really just an example of regeneration – restoring the whole body from 1 cell (fertilized egg). No wonder that regenerative repair, which can make up for disruption of pattern with flexible corrective processes, also occurs in embryogenesis. While not surprising given regulative development as a whole, it sheds new light on the definition of “master regulator” or “determinant” genes for specific developmental outcomes. While loss- and gain-of-function tests (such as those that had been performed for Nodal in the early years of the study of the LR pathway) may indicate that a certain gene’s expression is a driver of what happens next, it has to be kept in mind that this may not be the whole story. Two things can be readily missed by experiments that are narrowly focused on direct functional change of the gene expression and an immediate readout of direct downstream response genes. First, the downstream consequences could be wiped out by corrective mechanisms, which can limit the validity of “necessary and sufficient” claims for specific gene products with respect to final patterning outcome. Second, the results may be quite different if that gene’s expression is deranged by targeting much earlier steps (giving the organism time to notice the problem and activate robust repair pathways).
These data and analyses suggest a new direction in this field, focused on a systems-level understanding of how distinct molecular steps are multiplexed by the embryo for monitoring, setting, and continuously re-setting the LR identity of its tissues. In the sense that embryogenesis is an example of the more general phenomenon of regeneration (rebuilding the whole body from 1 cell), the LR asymmetry mechanisms may be a window on a much more general and widely-relevant phenomenon than just the patterning of the LR axis. The study of these dynamics could reveal fundamental aspects of how genetics interfaces with physics to implement robust, self-correcting systems. The impact of this would extend beyond development and birth defects, to the understanding of evolution, regenerative medicine, and artificial life.
Materials and Methods
Cloning
Subcloning was carried out into pCS2+ using standard methods as described previously [112].
Animal Husbandry
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and Tufts IACUC protocol #M2014-79. Xenopus embryos were collected and maintained according to standard protocols [252] in 0.1 x Modified Marc’s Ringers (MMR), pH 7.8, and staged according to [239].
Microinjection
Capped, synthetic mRNAs were dissolved in water and injected into embryos in 3% Ficoll using standard methods [252]. mRNA injections were made into the animal pole of eggs within 30 minutes post fertilization (mpf) at 14 \textdegree C, or into 1 of 2 cells of Stage 2 [239] embryos (as indicated) using borosilicate glass needles calibrated to deliver a 10 nl injection volume.
Laterality Assays
Xenopus embryos were analyzed for position (situs) of three organs; the heart, stomach and gallbladder [186] at Stage 45 [239]. Heterotaxic embryos were defined as having a reversal in one or more organs. Only embryos with normal dorsoanterior development and clear left- or right-sided organs were scored. A $\chi^2$ test was used to compare absolute counts of heterotaxic embryos.
In situ hybridization
Whole mount in situ hybridization was optimized using standard protocols [253,254] with probes against Xnr1 (the Xenopus Nodal) [221], Lefty [255] and Pitx2 [222] generated in vitro from linearized template using DIG labeling mix (Roche). A $\chi^2$ test was used to compare absolute counts of embryos with correct (expression on the left lateral plate mesoderm) versus incorrect (absent, bilateral or right-sided) marker expression.
Acknowledgements
We thank Joan Lemire and Jean-Francois Paré for cloning assistance and helpful comments on the manuscript, and other members of the Levin lab and the LR asymmetry and regeneration communities for many helpful discussions. We gratefully acknowledge support of the Templeton World Charity Foundation TWCF0089/AB55, sub award from Physical Science Oncology Center supported by Award Number U54CA143876 from the National Cancer Institute, and the Paul G. Allen Family Foundation.
Supplementary figures
- Supplementary Table 1: Experimental evidence for fixing grouped according to functional targets.
References
- C Neville. Animal asymmetry. Edward Arnold, illustrated edition, 1976.
- B Casey and B P Hackett. Left-right axis malformations in man and mouse. Current Opinion in Genetics & Development, 10(3):257–261, Jun 2000.
- M S Cohen, R H Anderson, M I Cohen, A M Atz, M Fogel, P J Gruber, L Lopez, J J Rome, and P M Weinberg. Controversies, genetics, diagnostic assessment, and outcomes relating to the heterotaxy syndrome. Cardiology in the Young, 17 Suppl 2:29–43, Sep 2007.
- B P Hackett. Formation and malformation of the vertebrate left-right axis. Current Molecular Medicine, 2(1):39–66, Feb 2002.
- H Peeters and K Devriendt. Human laterality disorders. European Journal of Medical Genetics, 49(5):349–362, Oct 2006.
- A F Ramsdell. Left-right asymmetry and congenital cardiac defects: getting to the heart of the matter in vertebrate left-right axis determination. Developmental Biology, 288(1):1–20, Dec 2005.
- C C LITTLE and H MCDONALD. Abnormalities of the mammae in the house mouse. Journal of Heredity, Jan 1945.
- T Nogi, Y E Yuan, D Sorocco, R Perez-Tomas, and M Levin. Eye regeneration assay reveals an invariant functional left-right asymmetry in the early bilaterian, dugesia japonica. Laterality, 10(3):193–205, May 2005.
- V P Pai, L N Vandenberg, D Blackiston, and M Levin. Neurally derived tissues in xenopus laevis embryos exhibit a consistent bioelectrical left-right asymmetry. Stem cells international, 2012:353491, Dec 2012.
- L J Paulozzi and J M Lary. Laterality patterns in infants with external birth defects. Teratology, 60(5):265–271, Nov 1999.
- H Schmidt. Supernumerary nipples: prevalence, size, sex and side predilection – a prospective clinical study. European Journal of Pediatrics, 157(10):821–823, Oct 1998.
- N E Breslow, N F Palmer, L R Hill, J Buring, and G J D’Angio. Wilms’ tumor: prognostic factors for patients without metastases at diagnosis: results of the national wilms' tumor study. Cancer, 41(4):1577–1589, Apr 1978.
- X Bustamante-Marín, J A Garness, and B Capel. Testicular teratomas: an intersection of pluripotency, differentiation and cancer biology. The International Journal of Developmental Biology, 57(2-4):201–210, 2013.
- J W Fuseler, J P Robichaux, H I Atiyah, and A F Ramsdell. Morphometric and fractal dimension analysis identifies early neoplastic changes in mammary epithelium of mmtv-cneu mice. Anticancer Research, 34(3):1171–1177, Mar 2014.
- J P Robichaux, R M Hallett, J W Fuseler, J A Hassell, and A F Ramsdell. Mammary glands exhibit molecular laterality and undergo left-right asymmetric ductal epithelial growth in mmtv-cneu mice. Oncogene, 34(15):2003–2010, Apr 2015.
- T A Sandson, P Y Wen, and M LeMay. Reversed cerebral asymmetry in women with breast cancer. The Lancet, 339(8792):523–524, Feb 1992.
- J M Veltmaat, A F Ramsdell, and E Sterneck. Positional variations in mammary gland development and cancer. Journal of mammary gland biology and neoplasia, 18(2):179–188, Jun 2013.
- K J Meador, D W Loring, P G Ray, S W Helman, B R Vazquez, and P J Neveu. Role of cerebral lateralization in control of immune processes in humans. Annals of Neurology, 55(6):840–844, Jun 2004.
- Y Q Shen, G Hebert, Y Su, E Moze, P J Neveu, and K S Li. In mice, production of plasma il-1 and il-6 in response to mptp is related to behavioral lateralization. Brain Research, 1045(1-2):31–37, May 2005.
- C Balaratnasingam, W H Morgan, V Johnstone, S J Cringle, and D Y Yu. Heterogeneous distribution of axonal cytoskeleton proteins in the human optic nerve. Investigative Ophthalmology & Visual Science, 50(6):2824–2838, Jun 2009.
- W Ludwig, 1932.
- C W Dunn. Complex colony-level organization of the deep-sea siphonophore bargmannia elongata (cnidaria, hydrozoa) is directionally asymmetric and arises by the subdivision of pro-buds. Developmental Dynamics, 234(4):835–845, Dec 2005.
- M Gardner. The new ambidextrous universe. Dover Publications, 3rd rev. ed. edition, 1990.
- C S Wu, E Ambler, R W Hayward, D D Hoppes, and R P Hudson. Experimental test of parity conservation in beta decay. Physical Review, 105(4):1413–1415, Feb 1957.
- V V Alpatov. Specific action of optical isomers of mepacrine upon dextral and sinistral strains of bacillus mycoides flügge. Nature, 158(4023):838, Dec 1946.
- P Gray, M L Dodds, and H Worthing. The effect of carbohydrates and inhibitors on heterotaxia in chick embryos. Anatomical Record, 78:77–78, 1940.
- V B Kasinov. The action of arginine, asparagine and atebrine stereomers upon the left and rightlemna minor plants. Biologia plantarum, 22(5):321–326, Sep 1980.
- D C Rife. Genetic studies of monozygotic twins, iii: mirror-imaging. The Journal of Heredity, 24(11):443–6, 1933.
- A E Boycott, C Diver, S L Garstang, and F M Turner. The inheritance of sinistrality in limnaea peregra (mollusca, pulmonata). Philosophical Transactions of the Royal Society B: Biological Sciences, 219(462-467):51–131, Jan 1931.
- D C Rife. Handedness, with special reference to twins. Genetics, 25(2):178–186, Mar 1940.
- J P Golding, T A Partridge, J R Beauchamp, T King, N A Brown, M Gassmann, and P S Zammit. Mouse myotomes pairs exhibit left-right asymmetric expression of mlc3f and alpha-skeletal actin. Developmental Dynamics, 231(4):795–800, Dec 2004.
- R C Alexander, N Breslin, C Molnar, J Richter, and S Mukherjee. Counter clockwise scalp hair whorl in schizophrenia. Biological Psychiatry, 32(9):842–845, Nov 1992.
- A A Beaton and G Mellor. Direction of hair whorl and handedness. Laterality, 12(4):295–301, Jul 2007.
- J S Hatfield. The genetic basis of hair whorl, handedness, and other phenotypes. Medical Hypotheses, 66(4):708–714, 2006.
- A Jansen, H Lohmann, S Scharfe, C Sehlmeyer, M Deppe, and S Knecht. The association between scalp hair-whorl direction, handedness and hemispheric language dominance: is there a common genetic basis of lateralization? Neuroimage, 35(2):853–861, Apr 2007.
- D W Smith and B T Gong. Scalp-hair patterning: its origin and significance relative to early brain and upper facial development. Teratology, 9(1):17–34, Feb 1974.
- R J Goss. Is antler asymmetry in reindeer and caribou genetically determined? Proceedings of the Reindeer/Caribou Symposium, 2:364–372, 1980.
- P J Neveu. Brain lateralization and immunomodulation. The International journal of neuroscience, 70(1-2):135–143, May 1993.
- P J Neveu. Cerebral lateralization and the immune system. International Review of Neurobiology, 52:303–323, 2002.
- S L Wise, K J Meador, W O Thompson, S S Avery, D W Loring, and B B Wray. Cerebral lateralization and histamine skin test asymmetries in humans. Annals of Allergy, 70(4):328–332, Apr 1993.
- S Dane, B Borekci, and S Kadanali. Right-sided lateralisation of ovarian cancer and right bias asymmetry for involved pelvic lymph nodes by ovarian cancer cells. Laterality, 13(5):393–402, Sep 2008.
- A J Klar. Breast cancer predisposition and brain hemispheric laterality specification likely share a common genetic cause. Breast disease, 33(1):49–52, 2011.
- J Wilting and M Hagedorn. Left-right asymmetry in embryonic development and breast cancer: common molecular determinants? Current Medicinal Chemistry, 18(36):5519–5527, 2011.
- M Fujinaga, J M Baden, T H Shepard, and R I Mazze. Nitrous oxide alters body laterality in rats. Teratology, 41(2):131–135, Feb 1990.
- J D Sedar. The influence of direct current fields upon the developmental pattern of the chick embryo. Journal of Experimental Zoology, 133(1):47–71, Oct 1956.
- R E Shenefelt. Morphogenesis of malformations in hamsters caused by retinoic acid: relation to dose and stage at treatment. teratology 5:103-18. 1972. Birth Defects Research. Part A, Clinical and Molecular Teratology, 88(10):847–862, Oct 2010.
- A S Trulioff, Y B Malashichev, and A S Ermakov. Artificial inversion of the left–right visceral asymmetry in vertebrates: Conceptual approaches and experimental solutions. Russian journal of developmental biology, 46(6):307–325, Nov 2015.
- H J Yost. Inhibition of proteoglycan synthesis eliminates left-right asymmetry in xenopus laevis cardiac looping. Development, 110(3):865–874, Nov 1990.
- M Levin, R L Johnson, C D Stern, M Kuehn, and C Tabin. A molecular pathway determining left-right asymmetry in chick embryogenesis. Cell, 82(5):803–814, Sep 1995.
- M Levin. Molecular basis of left-right asymmetry in chick development. PhD thesis, Genetics Department, Harvard University, 1996.
- M Levin. Left-right asymmetry in vertebrate embryogenesis. Bioessays: News and Reviews in Molecular, Cellular and Developmental Biology, 19(4):287–296, Apr 1997.
- M Levin, D J Roberts, L B Holmes, and C Tabin. Laterality defects in conjoined twins. Nature, 384(6607):321, Nov 1996.
- M Levin. Left–right asymmetry and the chick embryo. Seminars in cell & developmental biology, 9(1):67–76, Feb 1998.
- A F Ramsdell and H J Yost. Molecular mechanisms of vertebrate left-right development. Trends in Genetics, 14(11):459–465, Nov 1998.
- A Raya and J C Izpisúa Belmonte. Sequential transfer of left-right information during vertebrate embryo development. Current Opinion in Genetics & Development, 14(5):575–581, Oct 2004.
- R D Burdine and A F Schier. Conserved and divergent mechanisms in left-right axis formation. Genes & Development, 14(7):763–776, Apr 2000.
- M Mercola and M Levin. Left-right asymmetry determination in vertebrates. Annual Review of Cell and Developmental Biology, 17:779–805, 2001.
- T Nakamura and H Hamada. Left-right patterning: conserved and divergent mechanisms. Development, 139(18):3257–3262, Sep 2012.
- A Raya and J C Izpisua Belmonte. Unveiling the establishment of left-right asymmetry in the chick embryo. Mechanisms of Development, 121(9):1043–1054, Sep 2004.
- T Boettger, L Wittler, and M Kessel. Fgf8 functions in the specification of the right body side of the chick. Current Biology, 9(5):277–280, Mar 1999.
- A V Borodulin, F M Eroshkin, A V Bayramov, and A G Zaraisky. Noggin4 expression during chick embryonic development. The International Journal of Developmental Biology, 56(5):403–406, 2012.
- K Carneiro, C Donnet, T Rejtar, B L Karger, G A Barisone, E Díaz, S Kortagere, J M Lemire, and M Levin. Histone deacetylase activity is necessary for left-right patterning during vertebrate development. BMC Developmental Biology, 11:29, May 2011.
- M B Furtado, M J Solloway, V J Jones, M W Costa, C Biben, O Wolstein, J I Preis, D B Sparrow, Y Saga, S L Dunwoodie, and et al. Bmp/smad1 signaling sets a threshold for the left/right pathway in lateral plate mesoderm and limits availability of smad4. Genes & Development, 22(21):3037–3049, Nov 2008.
- R P Harvey. Links in the left/right axial pathway. Cell, 94(3):273–276, Aug 1998.
- M Levin, S Pagan, D J Roberts, J Cooke, M R Kuehn, and C J Tabin. Left/right patterning signals and the independent regulation of different aspects of situs in the chick embryo. Developmental Biology, 189(1):57–67, Sep 1997.
- M Levin. The roles of activin and follistatin signaling in chick gastrulation. The International Journal of Developmental Biology, 42(4):553–559, May 1998.
- S Marques, A C Borges, A C Silva, S Freitas, M Cordenonsi, and J A Belo. The activity of the nodal antagonist cerl-2 in the mouse node is required for correct l/r body axis. Genes & Development, 18(19):2342–2347, Oct 2004.
- E N Meyers and G R Martin. Differences in left-right axis pathways in mouse and chick: functions of fgf8 and shh. Science (New York), 285(5426):403–406, Jul 1999.
- A Schweickert, P Vick, M Getwan, T Weber, I Schneider, M Eberhardt, T Beyer, A Pachur, and M Blum. The nodal inhibitor coco is a critical target of leftward flow in xenopus. Current Biology, 20(8):738–743, Apr 2010.
- A Vonica and A H Brivanlou. The left-right axis is regulated by the interplay of coco, xnr1 and derrière in xenopus embryos. Developmental Biology, 303(1):281–294, Mar 2007.
- M Albrieux and M Villaz. Bilateral asymmetry of the inositol trisphosphate-mediated calcium signaling in two-cell ascidian embryos. Biology of the Cell, 92(3-4):277–284, Jul 2000.
- B Basu and M Brueckner. Cilia multifunctional organelles at the center of vertebrate left-right asymmetry. Current Topics in Developmental Biology, 85:151–174, 2008.
- S L Bauer Huang, Y Saheki, M K VanHoven, I Torayama, T Ishihara, I Katsura, A van der Linden, P Sengupta, and C I Bargmann. Left-right olfactory asymmetry results from antagonistic functions of voltage-activated calcium channels and the raw repeat protein olrn-1 in c. elegans. Neural Development, 2:24, Nov 2007.
- C Chang, Y W Hsieh, B J Lesch, C I Bargmann, and C F Chuang. Microtubule-based localization of a synaptic calcium-signaling complex is required for left-right neuronal asymmetry in c. elegans. Development, 138(16):3509–3518, Aug 2011.
- M Delling, A A Indzhykulian, X Liu, Y Li, T Xie, D P Corey, and D E Clapham. Primary cilia are not calcium-responsive mechanosensors. Nature, Mar 2016.
- J A Kreiling, Z L Balantac, A R Crawford, Y Ren, J Toure, S Zchut, L Kochilas, and R Creton. Suppression of the endoplasmic reticulum calcium pump during zebrafish gastrulation affects left-right asymmetry of the heart and brain. Mechanisms of Development, 125(5-6):396–410, Jun 2008.
- D P Norris. Cilia, calcium and the basis of left-right asymmetry. BMC Biology, 10:102, Dec 2012.
- A Raya, Y Kawakami, C Rodríguez-Esteban, M Ibañes, D Rasskin-Gutman, J Rodríguez-León, D Büscher, J A Feijó, and J C Izpisúa Belmonte. Notch activity acts as a sensor for extracellular calcium during vertebrate left-right determination. Nature, 427(6970):121–128, Jan 2004.
- G C Vilhais-Neto, M Maruhashi, K T Smith, M Vasseur-Cognet, A S Peterson, J L Workman, and O Pourquié. Rere controls retinoic acid signalling and somite bilateral symmetry. Nature, 463(7283):953–957, Feb 2010.
- C L Henley, V Lebedev, and M Feigel’man. Possible mechanisms for initiating macroscopic left-right asymmetry in developing organisms, page 54–62. AIP, 2009.
- C L Henley. Possible origins of macroscopic left-right asymmetry in organisms. Journal of statistical physics, 148(4):741–775, Sep 2012.
- T H Chen, J J Hsu, X Zhao, C Guo, M N Wong, Y Huang, Z Li, A Garfinkel, C M Ho, Y Tintut, and et al. Left-right symmetry breaking in tissue morphogenesis via cytoskeletal mechanics. Circulation Research, 110(4):551–559, Feb 2012.
- J Frankel. Intracellular handedness in ciliates. Ciba Foundation symposium, 162:73–88; discussion 88, 1991.
- W Halfter, W Reckhaus, and S Kröger. Nondirected axonal growth on basal lamina from avian embryonic neural retina. The Journal of Neuroscience, 7(11):3712–3722, Nov 1987.
- A M Heacock and B W Agranoff. Clockwise growth of neurites from retinal explants. Science (New York), 198(4312):64–66, Oct 1977.
- N H Mendelson and S L Keener. Clockwise and counterclockwise pinwheel colony morphologies of bacillus subtilis are correlated with the helix hand of the strain. Journal of Bacteriology, 151(1):455–457, Jul 1982.
- E M Nelsen, J Frankel, and L M Jenkins. Non-genic inheritance of cellular handedness. Development, 105(3):447–456, Mar 1989.
- F J Segerer, F Thüroff, A Piera Alberola, E Frey, and J O Rädler. Emergence and persistence of collective cell migration on small circular micropatterns. Physical Review Letters, 114(22):228102, Jun 2015.
- Y H Tee, T Shemesh, V Thiagarajan, R F Hariadi, K L Anderson, C Page, N Volkmann, D Hanein, S Sivaramakrishnan, M M Kozlov, and et al. Cellular chirality arising from the self-organization of the actin cytoskeleton. Nature Cell Biology, 17(4):445–457, Apr 2015.
- J F Walsh, M E Manwaring, and P A Tresco. Directional neurite outgrowth is enhanced by engineered meningeal cell-coated substrates. Tissue Engineering, 11(7-8):1085–1094, Aug 2005.
- L Q Wan, K Ronaldson, M Park, G Taylor, Y Zhang, J M Gimble, and G Vunjak-Novakovic. Micropatterned mammalian cells exhibit phenotype-specific left-right asymmetry. Proceedings of the National Academy of Sciences of the United States of America, 108(30):12295–12300, Jul 2011.
- L Q Wan and G Vunjak-Novakovic. Micropatterning chiral morphogenesis. Communicative & Integrative Biology, 4(6):745–748, Nov 2011.
- J Xu, A Van Keymeulen, N M Wakida, P Carlton, M W Berns, and H R Bourne. Polarity reveals intrinsic cell chirality. Proceedings of the National Academy of Sciences of the United States of America, 104(22):9296–9300, May 2007.
- H Yamanaka and S Kondo. Rotating pigment cells exhibit an intrinsic chirality. Genes To Cells, 20(1):29–35, Jan 2015.
- W L Zeile, F Zhang, R B Dickinson, and D L Purich. Listeria’s right-handed helical rocket-tail trajectories: mechanistic implications for force generation in actin-based motility. Cell Motility and the Cytoskeleton, 60(2):121–128, Feb 2005.
- K Sato, T Hiraiwa, E Maekawa, A Isomura, T Shibata, and E Kuranaga. Left-right asymmetric cell intercalation drives directional collective cell movement in epithelial morphogenesis. Nature Communications, 6:10074, Dec 2015.
- A Tamada, S Kawase, F Murakami, and H Kamiguchi. Autonomous right-screw rotation of growth cone filopodia drives neurite turning. The Journal of Cell Biology, 188(3):429–441, Feb 2010.
- Y Komatsu and Y Mishina. Establishment of left-right asymmetry in vertebrate development: the node in mouse embryos. Cellular and Molecular Life Sciences, 70(24):4659–4666, Dec 2013.
- J Schlueter and T Brand. Left-right axis development: examples of similar and divergent strategies to generate asymmetric morphogenesis in chick and mouse embryos. Cytogenetic and Genome Research, 117(1-4):256–267, 2007.
- M Levin and A R Palmer. Left-right patterning from the inside out: widespread evidence for intracellular control. Bioessays: News and Reviews in Molecular, Cellular and Developmental Biology, 29(3):271–287, Mar 2007.
- C Tabin. Do we know anything about how left-right asymmetry is first established in the vertebrate embryo? Journal of Molecular Histology, 36(5):317–323, Jun 2005.
- L N Vandenberg and M Levin. Perspectives and open problems in the early phases of left-right patterning. Seminars in Cell & Developmental Biology, 20(4):456–463, Jun 2009.
- L N Vandenberg and M Levin. Far from solved: a perspective on what we know about early mechanisms of left-right asymmetry. Developmental Dynamics, 239(12):3131–3146, Dec 2010.
- L N Vandenberg and M Levin. A unified model for left-right asymmetry? comparison and synthesis of molecular models of embryonic laterality. Developmental Biology, 379(1):1–15, Jul 2013.
- A Davison, G S McDowell, J M Holden, H F Johnson, G D Koutsovoulos, M M Liu, P Hulpiau, F Van Roy, C M Wade, R Banerjee, and et al. Formin is associated with left-right asymmetry in the pond snail and the frog. Current Biology, 26(5):654–660, Mar 2016.
- A Dimonte, A Adamatzky, V Erokhin, and M Levin. On chirality of slime mould. Bio Systems, 140:23–27, Feb 2016.
- R L Gardner. Normal bias in the direction of fetal rotation depends on blastomere composition during early cleavage in the mouse. Plos One, 5(3):e9610, Mar 2010.
- J Gros, K Feistel, C Viebahn, M Blum, and C J Tabin. Cell movements at hensen’s node establish left/right asymmetric gene expression in the chick. Science (New York), 324(5929):941–944, May 2009.
- A Gruhl and B Okamura. Development and myogenesis of the vermiform buddenbrockia (myxozoa) and implications for cnidarian body plan evolution. EvoDevo, 3(1):10, May 2012.
- M Levin, T Thorlin, K R Robinson, T Nogi, and M Mercola. Asymmetries in h+/k+-atpase and cell membrane potentials comprise a very early step in left-right patterning. Cell, 111(1):77–89, Oct 2002.
- M Lobikin, G Wang, J Xu, Y W Hsieh, C F Chuang, J M Lemire, and M Levin. Early, nonciliary role for microtubule proteins in left-right patterning is conserved across kingdoms. Proceedings of the National Academy of Sciences of the United States of America, 109(31):12586–12591, Jul 2012.
- G S McDowell, J M Lemire, J F Paré, G Cammarata, L A Lowery, and M Levin. Conserved roles for cytoskeletal components in determining laterality. Integrative Biology: Quantitative Biosciences from Nano to Macro, Feb 2016.
- S R Naganathan, S Fürthauer, M Nishikawa, F Jülicher, and S W Grill. Active torque generation by the actomyosin cell cortex drives left-right symmetry breaking. eLife, 3:e04165, Dec 2014.
- R M Onjiko, S E Morris, S A Moody, and P Nemes. Single-cell mass spectrometry with multi-solvent extraction identifies metabolic differences between left and right blastomeres in the 8-cell frog (xenopus) embryo. The Analyst, Mar 2016.
- S Schonegg, A Hyman, and W Wood. Timing and mechanism of the initial cue establishing handed left-right asymmetry incaenorhabditis elegans embryos. Genesis, 52(6):572–580, Jun 2014.
- R M Roberts, M Katayama, S R Magnuson, M T Falduto, and K E Torres. Transcript profiling of individual twin blastomeres derived by splitting two-cell stage murine embryos. Biology of Reproduction, 84(3):487–494, Mar 2011.
- J H Sun, Y Zhang, B Y Yin, J X Li, G S Liu, W Xu, and S Tang. Differential expression of axin1, cdc25c and cdkn2d mrna in 2-cell stage mouse blastomeres. Zygote (Cambridge, England), 20(3):305–310, Aug 2012.
- F Bangs, N Antonio, P Thongnuek, M Welten, M G Davey, J Briscoe, and C Tickle. Generation of mice with functional inactivation of talpid3, a gene first identified in chicken. Development, 138(15):3261–3272, Aug 2011.
- J Männer. Does an equivalent of the “ventral node” exist in chick embryos? a scanning electron microscopic study. Anatomy and embryology, 203(6):481–490, Jun 2001.
- D S Adams, K R Robinson, T Fukumoto, S Yuan, R C Albertson, P Yelick, L Kuo, M McSweeney, and M Levin. Early, h+-v-atpase-dependent proton flux is necessary for consistent left-right patterning of non-mammalian vertebrates. Development, 133(9):1657–1671, May 2006.
- S Aw, D S Adams, D Qiu, and M Levin. H,k-atpase protein localization and kir4.1 function reveal concordance of three axes during early determination of left-right asymmetry. Mechanisms of Development, 125(3-4):353–372, Apr 2008.
- D Qiu, S M Cheng, L Wozniak, M McSweeney, E Perrone, and M Levin. Localization and loss-of-function implicates ciliary proteins in early, cytoplasmic roles in left-right asymmetry. Developmental Dynamics, 234(1):176–189, Sep 2005.
- B Ohkawara and C Niehrs. An atf2-based luciferase reporter to monitor non-canonical wnt signaling in xenopus embryos. Developmental Dynamics, 240(1):188–194, Jan 2011.
- N J Oviedo and M Levin. Gap junctions provide new links in left-right patterning. Cell, 129(4):645–647, May 2007.
- A Armakolas and A J Klar. Left-right dynein motor implicated in selective chromatid segregation in mouse cells. Science (New York), 315(5808):100–101, Jan 2007.
- A J Klar. A model for specification of the left-right axis in vertebrates. Trends in Genetics, 10(11):392–396, Nov 1994.
- A J Klar. Support for the selective chromatid segregation hypothesis advanced for the mechanism of left-right body axis development in mice. Breast disease, 29:47–56, 2008.
- S Sauer and A J Klar. Left-right symmetry breaking in mice by left-right dynein may occur via a biased chromatid segregation mechanism, without directly involving the nodal gene. Frontiers in oncology, 2:166, Nov 2012.
- A J Klar. Selective chromatid segregation mechanism proposed for the human split hand/foot malformation development by chromosome 2 translocations: A perspective. Developmental Biology, Oct 2015.
- C D Cota, P Bagher, P Pelc, C O Smith, C R Bodner, and T M Gunn. Mice with mutations in mahogunin ring finger-1 (mgrn1) exhibit abnormal patterning of the left-right axis. Developmental Dynamics, 235(12):3438–3447, Dec 2006.
- D Srivastava and O Chakrabarti. Mahogunin-mediated α-tubulin ubiquitination via noncanonical k6 linkage regulates microtubule stability and mitotic spindle orientation. Cell death & disease, 5:e1064, Feb 2014.
- N A Brown and L Wolpert. The development of handedness in left/right asymmetry. Development, 109(1):1–9, May 1990.
- M Levin and N Nascone. Two molecular models of initial left-right asymmetry generation. Medical Hypotheses, 49(5):429–435, Nov 1997.
- S R Naganathan, T C Middelkoop, S Fürthauer, and S W Grill. Actomyosin-driven left-right asymmetry: from molecular torques to chiral self organization. Current Opinion in Cell Biology, 38:24–30, Feb 2016.
- D C Bergmann, J R Crew, J M Kramer, and W B Wood. Cuticle chirality and body handedness in caenorhabditis elegans. Developmental genetics, 23(3):164–174, 1998.
- H Hutter and R Schnabel. Establishment of left-right asymmetry in the caenorhabditis elegans embryo: a multistep process involving a series of inductive events. Development, 121(10):3417–3424, Oct 1995.
- C Pohl and Z Bao. Chiral forces organize left-right patterning in c. elegans by uncoupling midline and anteroposterior axis. Developmental Cell, 19(3):402–412, Sep 2010.
- W B Wood and D Kershaw. Handed asymmetry, handedness reversal and mechanisms of cell fate determination in nematode embryos. Ciba Foundation symposium, 162:143–59; discussion 159, 1991.
- M Abe, H Takahashi, and R Kuroda. Spiral cleavages determine the left-right body plan by regulating nodal pathway in monomorphic gastropods, physa acuta. The International Journal of Developmental Biology, 58(6-7-8):513–520, 2014.
- R Kuroda, B Endo, M Abe, and M Shimizu. Chiral blastomere arrangement dictates zygotic left-right asymmetry pathway in snails. Nature, 462(7274):790–794, Dec 2009.
- V N Meshcheryakov and L V Beloussov. Asymmetrical rotations of blastomeres in early cleavage of gastropoda. Wilhelm Roux’s Archives of Developmental Biology, 177(3):193–203, 1975.
- M Oliverio, M C Digilio, P Versacci, B Dallapiccola, and B Marino. Shells and heart: are human laterality and chirality of snails controlled by the same maternal genes? American Journal of Medical Genetics. Part A, 152A(10):2419–2425, Oct 2010.
- J B Coutelis, A G Petzoldt, P Spéder, M Suzanne, and S Noselli. Left-right asymmetry in drosophila. Seminars in Cell & Developmental Biology, 19(3):252–262, Jun 2008.
- N González-Morales, C Géminard, G Lebreton, D Cerezo, J B Coutelis, and S Noselli. The atypical cadherin dachsous controls left-right asymmetry in drosophila. Developmental Cell, 33(6):675–689, Jun 2015.
- M Bornens. Centrosome composition and microtubule anchoring mechanisms. Current Opinion in Cell Biology, 14(1):25–34, Feb 2002.
- S Lacomble, S Vaughan, C Gadelha, M K Morphew, M K Shaw, J R McIntosh, and K Gull. Basal body movements orchestrate membrane organelle division and cell morphogenesis in trypanosoma brucei. Journal of Cell Science, 123(Pt 17):2884–2891, Sep 2010.
- J Hagmann. Pattern formation and handedness in the cytoskeleton of human platelets. Proceedings of the National Academy of Sciences of the United States of America, 90(8):3280–3283, Apr 1993.
- S Thitamadee, K Tuchihara, and T Hashimoto. Microtubule basis for left-handed helical growth in arabidopsis. Nature, 417(6885):193–196, May 2002.
- D C Bergmann, M Lee, B Robertson, M F Tsou, L S Rose, and W B Wood. Embryonic handedness choice in c. elegans involves the galpha protein gpa-16. Development, 130(23):5731–5740, Dec 2003.
- D Bienkowska and C R Cowan. Centrosomes can initiate a polarity axis from any position within one-cell c. elegans embryos. Current Biology, 22(7):583–589, Apr 2012.
- M Bornens. The centrosome in cells and organisms. Science (New York), 335(6067):422–426, Jan 2012.
- C Géminard, N González-Morales, J B Coutelis, and S Noselli. The myosin id pathway and left-right asymmetry in drosophila. Genesis, 52(6):471–480, Jun 2014.
- R Hatori, T Ando, T Sasamura, N Nakazawa, M Nakamura, K Taniguchi, S Hozumi, J Kikuta, M Ishii, and K Matsuno. Left-right asymmetry is formed in individual cells by intrinsic cell chirality. Mechanisms of Development, 133:146–162, Aug 2014.
- S Hozumi, R Maeda, K Taniguchi, M Kanai, S Shirakabe, T Sasamura, P Spéder, S Noselli, T Aigaki, R Murakami, and et al. An unconventional myosin in drosophila reverses the default handedness in visceral organs. Nature, 440(7085):798–802, Apr 2006.
- M Nakamura, K Matsumoto, Y Iwamoto, T Muguruma, N Nakazawa, R Hatori, K Taniguchi, R Maeda, and K Matsuno. Reduced cell number in the hindgut epithelium disrupts hindgut left-right asymmetry in a mutant of pebble, encoding a rhogef, in drosophila embryos. Mechanisms of Development, 130(2-3):169–180, Feb 2013.
- T Okumura, T Sasamura, M Inatomi, S Hozumi, M Nakamura, R Hatori, K Taniguchi, N Nakazawa, E Suzuki, R Maeda, and et al. Class i myosins have overlapping and specialized functions in left-right asymmetric development in drosophila. Genetics, 199(4):1183–1199, Apr 2015.
- T Okumura, H Utsuno, J Kuroda, E Gittenberger, T Asami, and K Matsuno. The development and evolution of left-right asymmetry in invertebrates: lessons from drosophila and snails. Developmental Dynamics, 237(12):3497–3515, Dec 2008.
- A G Petzoldt, J B Coutelis, C Géminard, P Spéder, M Suzanne, D Cerezo, and S Noselli. De-cadherin regulates unconventional myosin id and myosin ic in drosophila left-right asymmetry establishment. Development, 139(10):1874–1884, May 2012.
- P Spéder, G Adám, and S Noselli. Type id unconventional myosin controls left-right asymmetry in drosophila. Nature, 440(7085):803–807, Apr 2006.
- K Taniguchi, R Maeda, T Ando, T Okumura, N Nakazawa, R Hatori, M Nakamura, S Hozumi, H Fujiwara, and K Matsuno. Chirality in planar cell shape contributes to left-right asymmetric epithelial morphogenesis. Science (New York), 333(6040):339–341, Jul 2011.
- S Pyrpassopoulos, E A Feeser, J N Mazerik, M J Tyska, and E M Ostap. Membrane-bound myo1c powers asymmetric motility of actin filaments. Current Biology, 22(18):1688–1692, Sep 2012.
- M Y Ali, S Uemura, K Adachi, H Itoh, K Kinosita, and S Ishiwata. Myosin v is a left-handed spiral motor on the right-handed actin helix. Nature Structural Biology, 9(6):464–467, Jun 2002.
- T Nishizaka, T Yagi, Y Tanaka, and S Ishiwata. Right-handed rotation of an actin filament in an in vitro motile system. Nature, 361(6409):269–271, Jan 1993.
- N A Brown, A McCarthy, and L Wolpert. Development of handed body asymmetry in mammals. Ciba Foundation symposium, 162:182–96; discussion 196, 1991.
- Y Han, A Lim, S Park, S Chang, S Lee, and W Ho. Rac-mediated actin remodeling and myosin ii are involved in katp channel trafficking in pancreatic β-cells. Experimental & Molecular Medicine, 47(10):e190, Oct 2015.
- P Müller, K W Rogers, B M Jordan, J S Lee, D Robson, S Ramanathan, and A F Schier. Differential diffusivity of nodal and lefty underlies a reaction-diffusion patterning system. Science (New York), 336(6082):721–724, May 2012.
- A F Schier. Nodal morphogens. Cold Spring Harbor Perspectives in Biology, 1(5):a003459, Nov 2009.
- N Bessodes, E Haillot, V Duboc, E Röttinger, F Lahaye, and T Lepage. Reciprocal signaling between the ectoderm and a mesendodermal left-right organizer directs left-right determination in the sea urchin embryo. PLoS Genetics, 8(12):e1003121, Dec 2012.
- T Fukumoto, R Blakely, and M Levin. Serotonin transporter function is an early step in left-right patterning in chick and frog embryos. Developmental Neuroscience, 27(6):349–363, 2005.
- T Fukumoto, I P Kema, and M Levin. Serotonin signaling is a very early step in patterning of the left-right axis in chick and frog embryos. Current Biology, 15(9):794–803, May 2005.
- A Garic-Stankovic, M Hernandez, G R Flentke, M H Zile, and S M Smith. A ryanodine receptordependent ca(i)(2+) asymmetry at hensen’s node mediates avian lateral identity. Development, 135(19):3271–3280, Oct 2008.
- A Raya, Y Kawakami, C Rodriguez-Esteban, D Buscher, C M Koth, T Itoh, M Morita, R M Raya, I Dubova, J G Bessa, and et al. Notch activity induces nodal expression and mediates the establishment of left-right asymmetry in vertebrate embryos. Genes & Development, 17(10):1213–1218, May 2003.
- L N Vandenberg, J M Lemire, and M Levin. Serotonin has early, cilia-independent roles in xenopus left-right patterning. Disease Models & Mechanisms, 6(1):261–268, Jan 2013.
- S Bertrand, D Aldea, S Oulion, L Subirana, A R de Lera, I Somorjai, and H Escriva. Evolution of the role of ra and fgf signals in the control of somitogenesis in chordates. Plos One, 10(9):e0136587, Sep 2015.
- V Duboc, E Röttinger, F Lapraz, L Besnardeau, and T Lepage. Left-right asymmetry in the sea urchin embryo is regulated by nodal signaling on the right side. Developmental Cell, 9(1):147–158, Jul 2005.
- T Hibino, Y Ishii, M Levin, and A Nishino. Ion flow regulates left-right asymmetry in sea urchin development. Development Genes and Evolution, 216(5):265–276, May 2006.
- A G Davies, J T Pierce-Shimomura, H Kim, M K VanHoven, T R Thiele, A Bonci, C I Bargmann, and S L McIntire. A central role of the bk potassium channel in behavioral responses to ethanol in c. elegans. Cell, 115(6):655–666, Dec 2003.
- A Sagasti, N Hisamoto, J Hyodo, M Tanaka-Hino, K Matsumoto, and C I Bargmann. The camkii unc-43 activates the mapkkk nsy-1 to execute a lateral signaling decision required for asymmetric olfactory neuron fates. Cell, 105(2):221–232, Apr 2001.
- E R Troemel, A Sagasti, and C I Bargmann. Lateral signaling mediated by axon contact and calcium entry regulates asymmetric odorant receptor expression in c. elegans. Cell, 99(4):387–398, Nov 1999.
- Y Kawakami, A Raya, R M Raya, C Rodríguez-Esteban, and J C Izpisúa Belmonte. Retinoic acid signalling links left-right asymmetric patterning and bilaterally symmetric somitogenesis in the zebrafish embryo. Nature, 435(7039):165–171, May 2005.
- A Navis, L Marjoram, and M Bagnat. Cftr controls lumen expansion and function of kupffer’s vesicle in zebrafish. Development, 140(8):1703–1712, Apr 2013.
- K A Fakhro, M Choi, S M Ware, J W Belmont, J A Towbin, R P Lifton, M K Khokha, and M Brueckner. Rare copy number variations in congenital heart disease patients identify unique genes in left-right patterning. Proceedings of the National Academy of Sciences of the United States of America, 108(7):2915–2920, Feb 2011.
- J R Kaltman, C Schramm, and G D Pearson. The national heart, lung, and blood institute bench to bassinet program: A new paradigm for translational research. Journal of the American College of Cardiology, 55(12):1262–1265, Mar 2010.
- R Bähring, H Standhardt, E A Martelli, and R Grantyn. Gaba-activated chloride currents of postnatal mouse retinal ganglion cells are blocked by acetylcholine and acetylcarnitine: How specific are ion channels in immature neurons? European Journal of Neuroscience, 6(7):1089–1099, Jul 1994.
- D Bowie and M L Mayer. Inward rectification of both ampa and kainate subtype glutamate receptors generated by polyamine-mediated ion channel block. Neuron, 15(2):453–462, Aug 1995.
- M Levin and M Mercola. Gap junctions are involved in the early generation of left-right asymmetry. Developmental Biology, 203(1):90–105, Nov 1998.
- R W Ransom and N L Stec. Cooperative modulation of [3h]mk-801 binding to the n-methyl-d-aspartate receptor-ion channel complex by l-glutamate, glycine, and polyamines. Journal of Neurochemistry, 51(3):830–836, Sep 1988.
- Y Sano, K Inamura, A Miyake, S Mochizuki, C Kitada, H Yokoi, K Nozawa, H Okada, H Matsushime, and K Furuichi. A novel two-pore domain k+ channel, tresk, is localized in the spinal cord. The Journal of Biological Chemistry, 278(30):27406–27412, Jul 2003.
- N C Spitzer. How gaba generates depolarization. The Journal of Physiology, 588(Pt 5):757–758, Mar 2010.
- M van der Stelt and V Di Marzo. Anandamide as an intracellular messenger regulating ion channel activity. Prostaglandins & Other Lipid Mediators, 77(1-4):111–122, Sep 2005.
- K Williams. Interactions of polyamines with ion channels. The Biochemical Journal, 325 ( Pt 2):289–297, Jul 1997.
- F Zhao, C P Song, J He, and H Zhu. Polyamines improve k+/na+ homeostasis in barley seedlings by regulating root ion channel activities. Plant Physiology, 145(3):1061–1072, Nov 2007.
- T Nakamura, N Mine, E Nakaguchi, A Mochizuki, M Yamamoto, K Yashiro, C Meno, and H Hamada. Generation of robust left-right asymmetry in the mouse embryo requires a self-enhancement and lateral-inhibition system. Developmental Cell, 11(4):495–504, Oct 2006.
- I C Welsh, M Thomsen, D W Gludish, C Alfonso-Parra, Y Bai, J F Martin, and N A Kurpios. Integration of left-right pitx2 transcription and wnt signaling drives asymmetric gut morphogenesis via daam2. Developmental Cell, 26(6):629–644, Sep 2013.
- C M Nelson and J P Gleghorn. Sculpting organs: mechanical regulation of tissue development. Annual Review of Biomedical Engineering, 14:129–154, Apr 2012.
- E S Noel, M Verhoeven, A K Lagendijk, F Tessadori, K Smith, S Choorapoikayil, J den Hertog, and J Bakkers. A nodal-independent and tissue-intrinsic mechanism controls heart-looping chirality. Nature Communications, 4:2754, 2013.
- J T Granados-Riveron and J D Brook. The impact of mechanical forces in heart morphogenesis. Circulation. Cardiovascular Genetics, 5(1):132–142, Feb 2012.
- S E Lindsey, J T Butcher, and H C Yalcin. Mechanical regulation of cardiac development. Frontiers in physiology, 5:318, Aug 2014.
- A Ramasubramanian, Q B Chu-Lagraff, T Buma, K T Chico, M E Carnes, K R Burnett, S A Bradner, and S S Gordon. On the role of intrinsic and extrinsic forces in early cardiac s-looping. Developmental Dynamics, 242(7):801–816, Jul 2013.
- D A Voronov, P W Alford, G Xu, and L A Taber. The role of mechanical forces in dextral rotation during cardiac looping in the chick embryo. Developmental Biology, 272(2):339–350, Aug 2004.
- J K Muller, D R Prather, and N M Nascone-Yoder. Left-right asymmetric morphogenesis in the xenopus digestive system. Developmental Dynamics, 228(4):672–682, Dec 2003.
- K S Latacha, M C Rémond, A Ramasubramanian, A Y Chen, E L Elson, and L A Taber. Role of actin polymerization in bending of the early heart tube. Developmental Dynamics, 233(4):1272–1286, Aug 2005.
- Y Shi, J Yao, J M Young, J A Fee, R Perucchio, and L A Taber. Bending and twisting the embryonic heart: a computational model for c-looping based on realistic geometry. Frontiers in physiology, 5:297, Aug 2014.
- T Tsuda, N Philp, M H Zile, and K K Linask. Left-right asymmetric localization of flectin in the extracellular matrix during heart looping. Developmental Biology, 173(1):39–50, Jan 1996.
- H Driesch. The science & philosophy of the organism. A. and C. Black, 1908.
- L N Vandenberg, D S Adams, and M Levin. Normalized shape and location of perturbed craniofacial structures in the xenopus tadpole reveal an innate ability to achieve correct morphology. Developmental Dynamics, 241(5):863–878, May 2012.
- A Eldar, R Dorfman, D Weiss, H Ashe, B Z Shilo, and N Barkai. Robustness of the bmp morphogen gradient in drosophila embryonic patterning. Nature, 419(6904):304–308, Sep 2002.
- A Eldar, B Z Shilo, and N Barkai. Elucidating mechanisms underlying robustness of morphogen gradients. Current Opinion in Genetics & Development, 14(4):435–439, Aug 2004.
- M Joachimczak and B Wróbel. Evolution of robustness to damage in artificial 3-dimensional development. Bio Systems, 109(3):498–505, Sep 2012.
- K D Birnbaum and A Sánchez Alvarado. Slicing across kingdoms: regeneration in plants and animals. Cell, 132(4):697–710, Feb 2008.
- S J Forbes and N Rosenthal. Preparing the ground for tissue regeneration: from mechanism to therapy. Nature Medicine, 20(8):857–869, Aug 2014.
- M Levin. Reprogramming cells and tissue patterning via bioelectrical pathways: molecular mechanisms and biomedical opportunities. Wiley interdisciplinary reviews. Systems biology and medicine, 5(6):657–676, Dec 2013.
- T D Bunney, A H De Boer, and M Levin. Fusicoccin signaling reveals 14-3-3 protein function as a novel step in left-right patterning during amphibian embryogenesis. Development, 130(20):4847–4858, Oct 2003.
- J L Lohr, M C Danos, and H J Yost. Left-right asymmetry of a nodal-related gene is regulated by dorsoanterior midline structures during xenopus development. Development, 124(8):1465–1472, Apr 1997.
- I F Tabry, J Calabrese, H Zammar, K Abou-Kasem, H Akeilan, N Gharbieh, H Zinati, W Noureddine, A el Hout, M Tayah, and et al. Case report: off-pump total myocardial revascularization for dextrocardia and situs inversus. The Heart Surgery Forum, 4(3):251–253, 2001.
- S Aw, J C Koster, W Pearson, C G Nichols, N Q Shi, K Carneiro, and M Levin. The atp-sensitive k(+)-channel (k(atp)) controls early left-right patterning in xenopus and chick embryos. Developmental Biology, 346(1):39–53, Oct 2010.
- B A Hyatt and H J Yost. The left-right coordinator: the role of vg1 in organizing left-right axis formation. Cell, 93(1):37–46, Apr 1998.
- B A Hyatt, J L Lohr, and H J Yost. Initiation of vertebrate left-right axis formation by maternal vg1. Nature, 384(6604):62–65, Nov 1996.
- J L Lohr, M C Danos, T W Groth, and H J Yost. Maintenance of asymmetric nodal expression in xenopus laevis. Developmental genetics, 23(3):194–202, 1998.
- A K Ryan, B Blumberg, C Rodriguez-Esteban, S Yonei-Tamura, K Tamura, T Tsukui, J de la Peña, W Sabbagh, J Greenwald, S Choe, and et al. Pitx2 determines left-right asymmetry of internal organs in vertebrates. Nature, 394(6693):545–551, Aug 1998.
- K Sampath, A M Cheng, A Frisch, and C V Wright. Functional differences among xenopus nodal-related genes in left-right axis determination. Development, 124(17):3293–3302, Sep 1997.
- M Campione, H Steinbeisser, A Schweickert, K Deissler, F van Bebber, L A Lowe, S Nowotschin, C Viebahn, P Haffter, M R Kuehn, and et al. The homeobox gene pitx2: mediator of asymmetric left-right signaling in vertebrate heart and gut looping. Development, 126(6):1225–1234, Mar 1999.
- J Beisson and T M Sonneborn. Cytoplasmic inheritance of the organization of the cell cortex in paramecium aurelia. Proceedings of the National Academy of Sciences of the United States of America, 53:275–282, Feb 1965.
- J Frankel. Positional information in unicellular organisms. Journal of Theoretical Biology, 47(2):439–481, Oct 1974.
- J Frankel. Positional order and cellular handedness. Journal of Cell Science, 97 ( Pt 2):205–211, Oct 1990.
- X B Shi, Z J Qiu, and J Frankel. Morphology and development of left-handed singlets derived from mirror-image doublets of stylonychia mytilus. The Journal of protozoology, 37(1):14–19, Feb 1990.
- V Levy and O Khaner. Limited left-right cell migration across the midline of the gastrulating avian embryo. Developmental genetics, 23(3):175–184, 1998.
- M Levin and M Mercola. Gap junction-mediated transfer of left-right patterning signals in the early chick blastoderm is upstream of shh asymmetry in the node. Development, 126(21):4703–4714, Nov 1999.
- M C Danos and H J Yost. Linkage of cardiac left-right asymmetry and dorsal-anterior development in xenopus. Development, 121(5):1467–1474, May 1995.
- K A Kelly, Y Wei, and T Mikawa. Cell death along the embryo midline regulates left-right sidedness. Developmental Dynamics, 224(2):238–244, Jun 2002.
- M L Martínez-Frías, M Urioste, E Bermejo, E Rodríguez-Pinilla, V Félix, L Paisán, S Martínez, J Egüés, F Gómez, and P Aparicio. Primary midline developmental field. ii. clinical/epidemiological analysis of alteration of laterality (normal body symmetry and asymmetry). American Journal of Medical Genetics, 56(4):382–388, May 1995.
- M L Martínez-Frías. Primary midline developmental field. i. clinical and epidemiological characteristics. American Journal of Medical Genetics, 56(4):374–381, May 1995.
- E Roessler and M Muenke. Midline and laterality defects: left and right meet in the middle. Bioessays: News and Reviews in Molecular, Cellular and Developmental Biology, 23(10):888–900, Oct 2001.
- R J Agate, W Grisham, J Wade, S Mann, J Wingfield, C Schanen, A Palotie, and A P Arnold. Neural, not gonadal, origin of brain sex differences in a gynandromorphic finch. Proceedings of the National Academy of Sciences of the United States of America, 100(8):4873–4878, Apr 2003.
- F B Hutt. Genetics of the Fowl: The Classic Guide to Chicken Genetics and Poultry Breeding. Norton Creek Press, paperback; 2003-05-01 edition, 2003.
- D Zhao, D McBride, S Nandi, H A McQueen, M J McGrew, P M Hocking, P D Lewis, H M Sang, and M Clinton. Somatic sex identity is cell autonomous in the chicken. Nature, 464(7286):237–242, Mar 2010.
- S Aw and M Levin. What’s left in asymmetry? Developmental Dynamics, 237(12):3453–3463, Dec 2008.
- L N Vandenberg, B W Pennarola, and M Levin. Low frequency vibrations disrupt left-right patterning in the xenopus embryo. Plos One, 6(8):e23306, Aug 2011.
- P D Nieuwkoop and J Faber. Normal Table of Xenopus Laevis. Garland Science, 1st edition, 1994.
- L N Vandenberg. Laterality defects are influenced by timing of treatments and animal model. Differentiation; Research in Biological Diversity, 83(1):26–37, Jan 2012.
- P Csermely, J Hódsági, T Korcsmáros, D Módos, Á R Perez-Lopez, K Szalay, D V Veres, K Lenti, L Y Wu, and X S Zhang. Cancer stem cells display extremely large evolvability: alternating plastic and rigid networks as a potential mechanism: network models, novel therapeutic target strategies, and the contributions of hypoxia, inflammation and cellular senescence. Seminars in Cancer Biology, 30:42–51, Feb 2015.
- P Csermely and T Korcsmáros. Cancer-related networks: a help to understand, predict and change malignant transformation. Seminars in Cancer Biology, 23(4):209–212, Aug 2013.
- S Huang, I Ernberg, and S Kauffman. Cancer attractors: a systems view of tumors from a gene network dynamics and developmental perspective. Seminars in Cell & Developmental Biology, 20(7):869–876, Sep 2009.
- S Huang and D E Ingber. A non-genetic basis for cancer progression and metastasis: self-organizing attractors in cell regulatory networks. Breast disease, 26:27–54.
- S Huang. On the intrinsic inevitability of cancer: from foetal to fatal attraction. Seminars in Cancer Biology, 21(3):183–199, Jun 2011.
- J G Zañudo and R Albert. Cell fate reprogramming by control of intracellular network dynamics. PLoS Computational Biology, 11(4):e1004193, Apr 2015.
- K Friston, M Levin, B Sengupta, and G Pezzulo. Knowing one’s place: a free-energy approach to pattern regulation. Journal of the Royal Society, Interface, 12(105), Apr 2015.
- R Gordon and R Stone. Cybernetic Embryo. World Scientific Publishing Co, 2016.
- H Hauser, A J Ijspeert, R M Füchslin, R Pfeifer, and W Maass. Towards a theoretical foundation for morphological computation with compliant bodies. Biological Cybernetics, Jan 2012.
- J Mustard and M Levin. Bioelectrical mechanisms for programming growth and form: Taming physiological networks for soft body robotics. Soft Robotics, 1(3):169–191, Sep 2014.
- G Pezzulo and M Levin. Re-membering the body: applications of computational neuroscience to the top-down control of regeneration of limbs and other complex organs. Integrative Biology: Quantitative Biosciences from Nano to Macro, 7(12):1487–1517, Dec 2015.
- H L Sive, R M Grainger, and R M Harland. Early Development of Xenopus Laevis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, 2000.
- R M Harland. In situ hybridization: an improved whole-mount method for xenopus embryos. Methods in Cell Biology, 36:685–695, 1991.
- A H Monsoro-Burq. A rapid protocol for whole-mount in situ hybridization on xenopus embryos. CSH protocols, 2007:pdb.prot4809, Aug 2007.
- C Meno, Y Ito, Y Saijoh, Y Matsuda, K Tashiro, S Kuhara, and H Hamada. Two closely-related left-right asymmetrically expressed genes, lefty-1 and lefty-2: their distinct expression domains, chromosomal linkage and direct neuralizing activity in xenopus embryos. Genes To Cells, 2(8):513–524, Aug 1997.